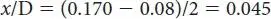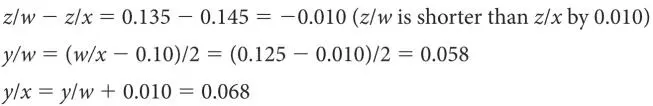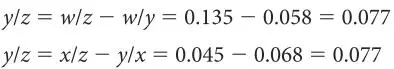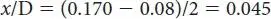
x /C is the same 0.045 plus the 0.08 by which we already determined it was longer = 0.125:
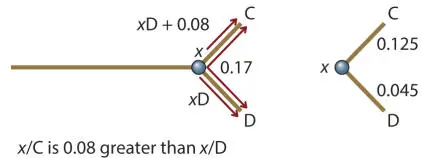
Branches z /E and z /F
The distance between E and F can be calculated in the same way:

The length of E/ z /F (E/F) is 0.30, and the average difference between anywhere else in the tree and E or F is −0.08. The negative sign means that the branch to F is longer than the branch to E. So z /E = (0.30 − 0.08)/2 = 0.11, and z /F − 0.11 + 0.08 = 0.1.
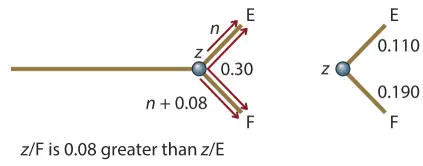
Internal branches w / x , x / y , and x / z
The tree so far is:
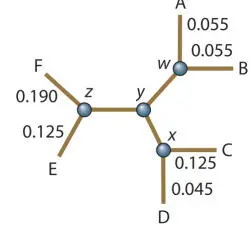
To solve the lengths of internal branches, the same process is used for collections of branches and then the average length outside the internal nodes is subtracted. To determine the lengths of w/y, x/y , and z/y , we need to determine the total lengths of w/x, x/z , and z/w and then figure out where along these to place y .
To solve for the length of w/x , the average distance between AB and CD in the reduced matrix (0.265) is used, and then the average w /A and w /B distance (AB/2 = 0.11/2 = 0.055) and the average x /C and x /D distance (CD/2 = 0.17/2 = 0.085) are subtracted to leave the w/x distance: 0.265 − 0.055 − 0.085 = 0.125.
To solve the length of x/z , the average CD/EF distance is calculated (this was not done in the reduced matrices, but it is the same process; it is 0.38) and then x /CD (CD/2 = 0.085) and x /EF (EF/2 = 0.30/2 = 0.150) are subtracted to give x/z = 0.145. Likewise, z/w = EF/AB (0.34) − w /AB (0.055) − z /EF (0.15) = 0.135.
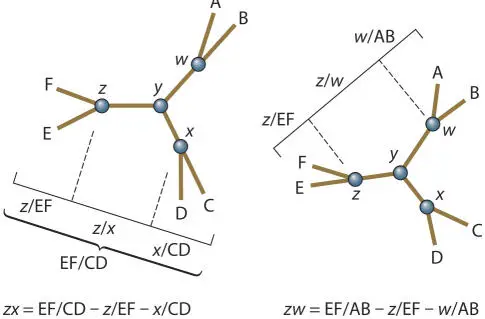
Thus, w/x = 0.125, x/z = 0.145, and z/w = 0.135.
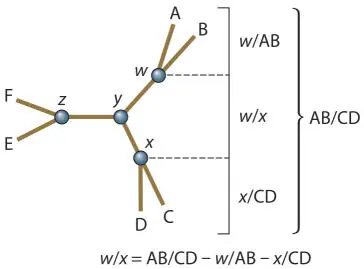
To determine where on any of these line segments node y is, we need to pick any of the line segments and calculate the difference to each end of this segment from the other branch. We do this just like we did for solving the lengths of the branches between A/B, C/D, or E/F. For example, for branch w/x :
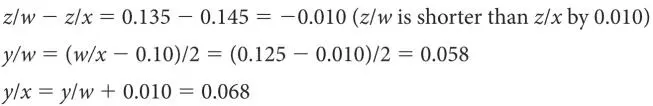
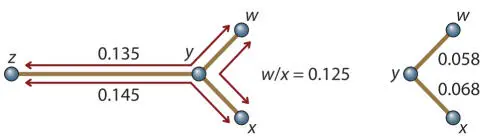
Now we know all of the line segment lengths but one, y/z . We can get this by subtracting all of the known lengths of w/y from w/z , or all the known lengths of x/y from x/z ; either way we get the same answer:
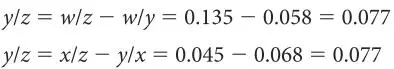

So, our final tree looks like this:
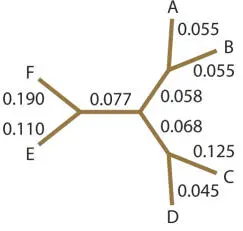
or like this with the branches drawn to scale:
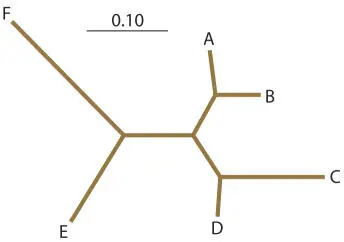
Notice that in the final tree, the evolutionary distance between any two sequences is approximately equal (any difference is due to the need to average branch lengths) to the sum of the lengths of the line segments joining those two sequences; in other words, the tree is additive. This should be true on any quantitative phylogenetic tree.
The neighbor-joining method is very fast and so can be used on trees with much larger numbers of sequences than can other methods, which are discussed in chapter 5.
Rooting a tree with an outgroup
Whenever possible, an outgroup sequence should be included in the analysis; an outgroup is a sequence that is known to be outside of the group you are interested in treeing. Only by including an outgroup can the root of a tree be located. For example, when building trees from mammalian sequences, the sequence from a reptile might be included as an outgroup. Outgroups provide the root to the rest of the tree; although no tree generated by these methods has a real root, if other information shows that one (or a set) of the sequences is unrelated to the rest, wherever that branch connects to the rest of the tree defines the root (common ancestor) of that portion of the tree. In our example tree above, sequences A to D might be mammalian whereas E and F might be reptilian. If the tree included only mammalian sequences, it would be impossible to know where the root is, but the inclusion of an outgroup provides that information.
Molecular phylogenetic trees depict the relationships between gene sequences
The final step in the tree-building process is the leap of faith that the tree depicting the relationships between the sequence similarities in an alignment also depicts the evolutionary history of the organisms from which the sequences were derived. You can go wrong for many reasons at this step.
The most common reason is the choice of sequence for use in the analysis, the first step of the whole process. What would happen if, for example, you made a tree of mammals by using globin genes, but used alpha globin sequences from some species and beta globin genes from others? What might happen if, in an analysis of plants, you used nuclear SSU rRNA sequences from some plants, chloroplast SSU rRNAs from others, and mitochondrial sequences from still others? What if, in an analysis of bacterial species, you used a gene like amp , encoding penicillin resistance, which clearly moves readily from species to species? The trees you would build with these sequences might be perfectly valid, accurately representing the relationships between the genes used, but they would not represent the relationships between the organisms.
Читать дальше