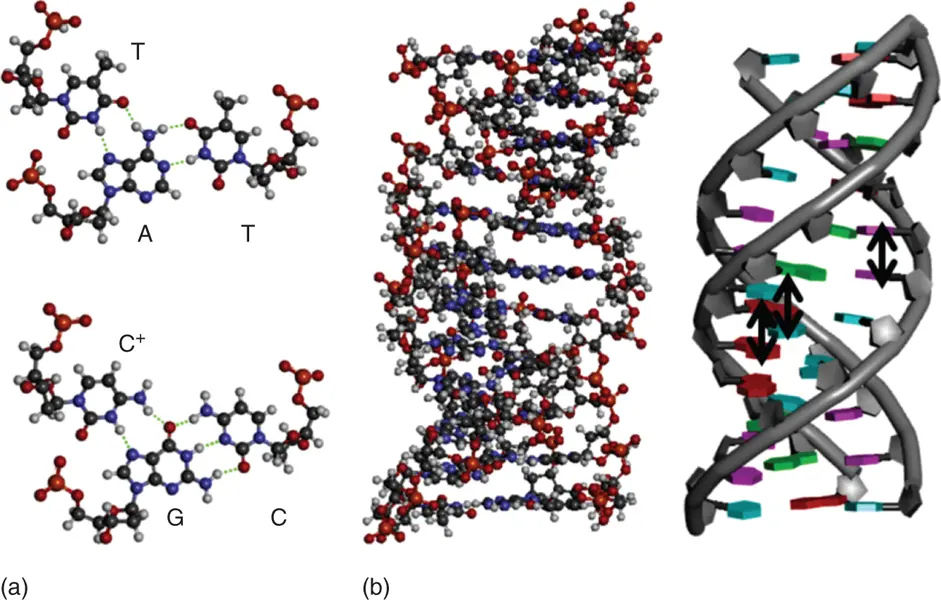
Figure 3.5 (a) Hydrogen bond formation in base triads in a DNA triplex. The hydrogen bonds are shown in dashed lines. (b) Triplex structures of DNAs are depicted in tube (left) and ball-and-stick (right) models, respectively. Stacking interactions are shown in arrows.
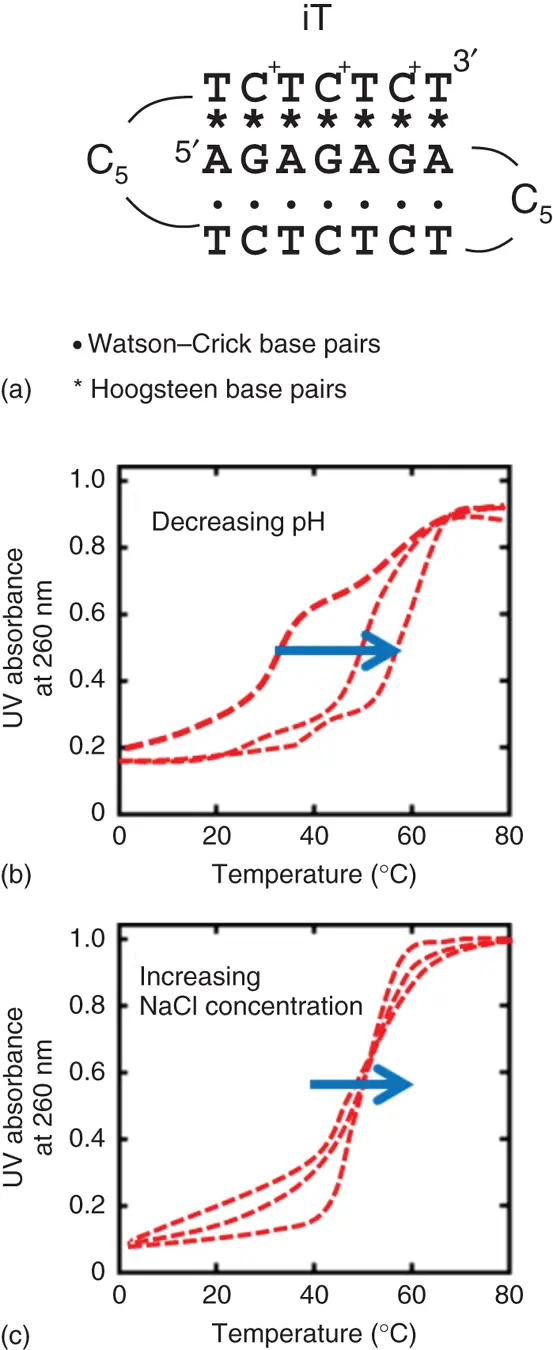
Figure 3.6 (a) Sequence for the intermolecular triplex (iT). Typical images of UV melting curves for iT in (b) various pH solutions or (c) various concentrations of Na +.
Moreover, the unfolding processes for the triplexes also depend on the cation concentration due to high charge density of triplexes ( Figure 3.6c). In general, the stabilities of triplexes are much greater with divalent cations than with monovalent cations, and almost a 100-fold concentration of Na +is required to stabilize the triplex as effectively as Mg 2+or Mn 2+[7]. It has been reported that Mg 2+is crucial for the formation of the Y-R*R triplexes than that for the formation of the Y-R*Y triplexes ( Figure 3.7) [8]. The protonation at N3 of Hoogsteen cytosine residues plays a key role in stabilizing the Y-R*Y triplex structure. In contrast, the Y-R*R triplex does not require this precondition; therefore, the interaction of divalent cations with a triplex may be different for the two types of triplexes. The interaction of metal ions with the triplexes clearly depended on the type and ionic strength of the cations, and the efficiency with which the cations stabilized the global triplex was in the order Mg 2+> Mn 2+> Ca 2+> Ba 2+> Na +(monovalent cations). This order is in good agreement with the ionic radii ( r ) of the divalent cations ( r of Mg 2+; 0.66 Å, r of Mn 2+; 0.80 Å, r of Ca 2+; 0.99 Å, r of Ba 2+; 1.34 Å), which suggests that the divalent cations with a smaller radius may increase the affinity of the nucleotide alignment, resulting in enhancing the stability of the double helix structure. It was reported that the folding of RNA tertiary structures is also stabilized by cations and the stabilizing ability of cations to RNA depends on the ionic radius [9].
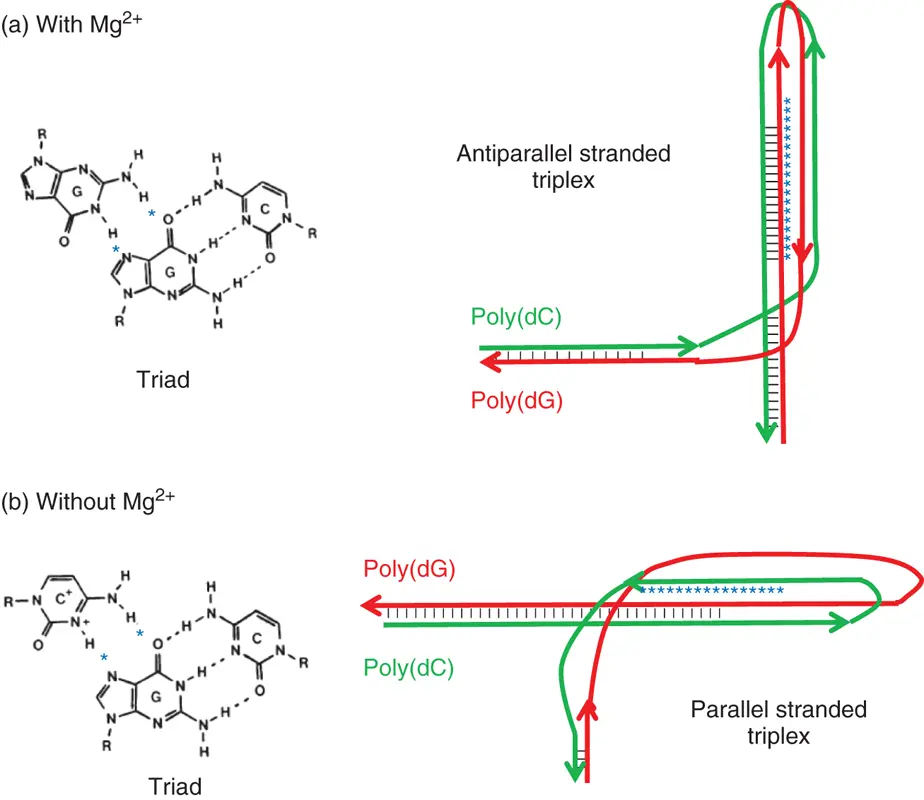
Figure 3.7 Probable structures adopted by the poly(dG)-poly(dC) sequence in supercoiled plasmid DNA and the corresponding hydrogen bonding scheme for the base triad in (a) the presence or (b) absence of 2 mM Mg 2+. Hydrogen bonds in the non-B-DNA structure are shown in stars.
3.4.2 Factors Influencing Stability of Quadruplex
3.4.2.1 G-Quadruplexes
The basic subunit of a G-quadruplex structure is the G-quartet ( Figure 3.8a), composed of a planar cyclic array of four Hoogsteen-paired guanine residues ( Figure 3.8aandb) [10]. The spatial proximity of the O6 atoms of guanine in a quartet is unique, with their lone pairs in close approach. This arrangement would be expected to produce significant electrostatic repulsion among the guanine bases. Therefore, the coordination of cations in the center of the G-quartet is required for the stable G-quadruplex formations. The direct cation coordination is investigated by the melting temperature ( T m) of guanosine gels, which is a model of G-quartet in the presence of cations ( Figure 3.9) [11]. The T mvalues strongly correlate with ionic radius. Moreover, it is also reported that the T mvalues for the G-quadruplexes in the presence of various cations show similar trend with the T mvalues for guanosine gels. The UV melting profiles for the thrombin aptamer G-quadruplexes show that cations of K +, Rb +, NH 4+, Sr 2+, and Ba 2+are able to form stable intramolecular cation–G-quadruplex complexes at temperatures above 25 °C. The cations of Li +, Na +, Cs +, Mg 2+, and Ca 2+form weaker complexes at very low temperatures (the detail parameters are described below).
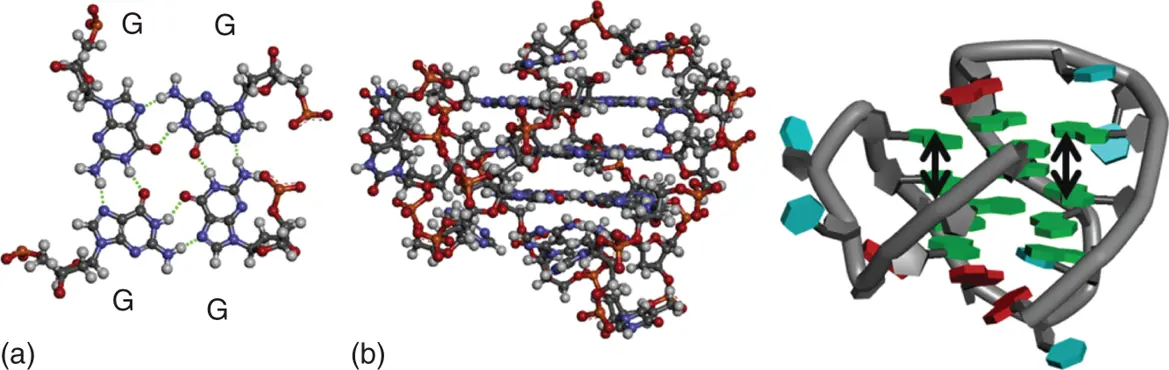
Figure 3.8 (a) Hydrogen bond formation in base pairs in a DNA G-quadruplex. The hydrogen bonds are shown in dashed lines. (b) G-quadruplex structures of DNAs are depicted in tube (left) and ball-and-stick (right) models, respectively. Stacking interactions are shown in arrows.
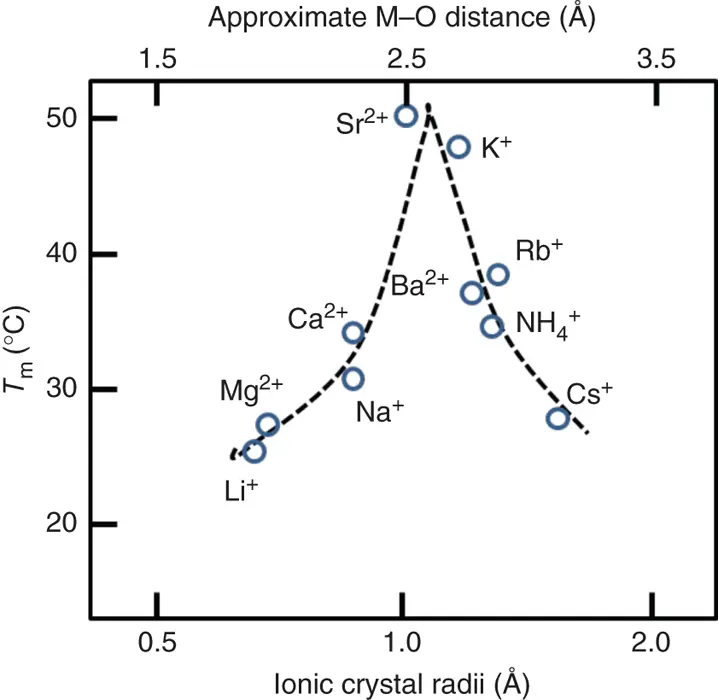
Figure 3.9 Images of the melting temperature of 8-bromoguanosine gels as a function of cation size.
The space among the four O6 carbonyl groups at each guanine in a G-quartet is sufficiently large to accommodate an Na +coordinated in the plane of the guanine bases. The ionic radius of Na +is 0.95 Å, and this size is around maximum size of a cation that can be coordinated in the plane of a G-quartet. Larger cations, such as K +, with an ionic radius of 1.33 Å, are too large to be coordinated within G-quartet. Importantly, larger cations are coordinated between the planes of two stacked G-quartets, and the coordination induces large stabilization of G-quadruplex. Cations with an ionic radius in the range of 1.3–1.5 Å fit well within the two G-quartets of the complex, while the other cations cannot. The cations coordinate to two stacked G-quartets with accompanying dehydration of both cations and guanine O6 atomic groups in a G-quartet, determining cation-dependent stability of G-quadruplexes.
C-rich sequences have been shown to form i-motif structures – a second type of quadruplex structure containing intercalated cytosine-protonated cytosine base pairs coupled together with looping base ( Figure 3.10a) [12]. For example, the i-motif DNA tetrameric structure is formed of two parallel duplexes intercalated in a head-to-tail orientation and held together by hemiprotonated cytosine pairs ( Figure 3.10b). The four phosphodiester backbones forming the structure define two narrow and wide grooves. The short interphosphate distances across the narrow groove and the charged C-imino protons induce a strong repulsion that should destabilize the i-motif. On the other hand, attractive interactions are observed between C-C pairs. Attractive interactions are also expected between C4–N4 and C2–O2 dipoles of stacked C-C pairs, which are nearly antiparallel in the i-motif structures, and between the N3 and H1 atoms of the cytosines forming a hemiprotonated C-C pair. The sum of all attractive interactions between the bases can thus compensate for the C-imino proton repulsion. Moreover, the twisting of the phosphodiester backbone is critical for the i-motif stability, and therefore all the phosphodiester backbones should display the same twist to produce a good stacking of bases. An i-motif structure of RNA is very unstable due to rigidity of backbones; therefore, the i-motif structures that are revealed by structural analysis such as NMR or X-ray analysis are composed of DNA.
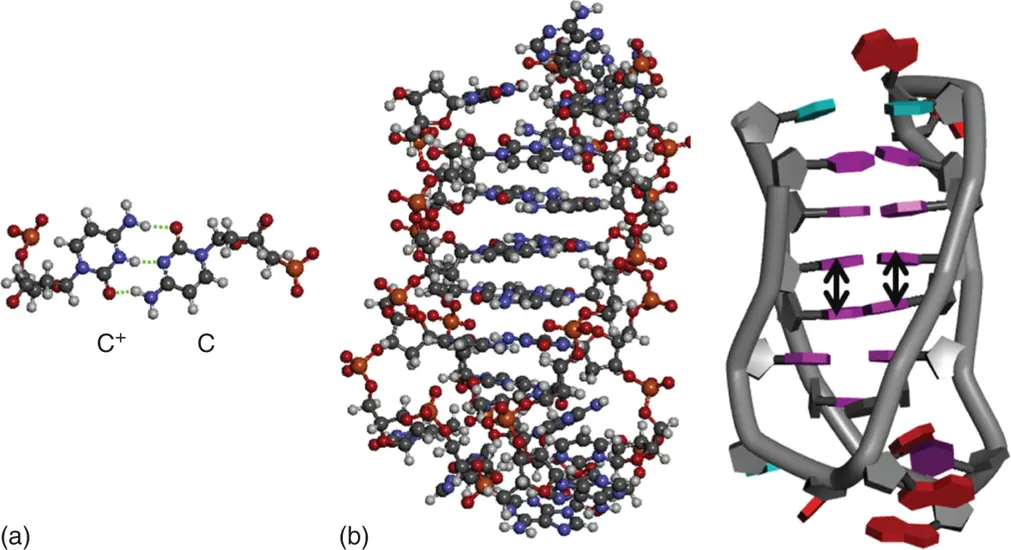
Figure 3.10 (a) Hydrogen bond formation in base pairs in a DNA i-motif. The hydrogen bonds are shown in dashed lines. (b) i-Motif structures of DNAs are depicted in tube (left) and ball-and-stick (right) models, respectively. Stacking interactions are shown in arrows.
Читать дальше


















