1 ...8 9 10 12 13 14 ...50 Nishimura and Suchard [57] turns CG into a viable algorithm for Bayesian sparse regression problems by realizing that (i) we can obtain a Gaussian vector 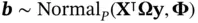 by first generating
by first generating 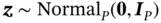 and
and 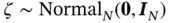 and then setting
and then setting 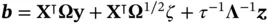 and (ii) subsequently solving
and (ii) subsequently solving 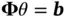 yields a sample
yields a sample 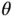 from the distribution ( 4). The authors then observe that the mechanism through which a shrinkage prior induces sparsity of
from the distribution ( 4). The authors then observe that the mechanism through which a shrinkage prior induces sparsity of 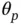 s also induces a tight clustering of eigenvalues in the prior‐preconditioned matrix
s also induces a tight clustering of eigenvalues in the prior‐preconditioned matrix 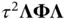 . This fact makes it possible for prior‐preconditioned CG to solve the system
. This fact makes it possible for prior‐preconditioned CG to solve the system  in
in 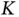 matrix–vector operations of form
matrix–vector operations of form 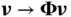 , where
, where 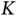 roughly represents the number of significant
roughly represents the number of significant 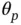 s that are distinguishable from zeros under the posterior. For
s that are distinguishable from zeros under the posterior. For 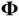 having a structure as in ( 4),
having a structure as in ( 4),  can be computed via matrix–vector multiplications of form
can be computed via matrix–vector multiplications of form 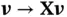 and
and 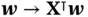 , so each
, so each 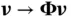 operation requires a fraction of the computational cost of directly computing
operation requires a fraction of the computational cost of directly computing  and then factorizing it.
and then factorizing it.
Prior‐preconditioned CG demonstrates an order of magnitude speedup in posterior computation when applied to a comparative effectiveness study of atrial fibrillation treatment involving 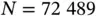 patients and
patients and 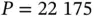 covariates [57]. Though unexplored in their work, the algorithm's heavy use of matrix–vector multiplications provides avenues for further acceleration. Technically, the algorithm's complexity may be characterized as
covariates [57]. Though unexplored in their work, the algorithm's heavy use of matrix–vector multiplications provides avenues for further acceleration. Technically, the algorithm's complexity may be characterized as 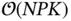 , for the
, for the 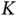 matrix–vector multiplications by
matrix–vector multiplications by 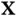 and
and 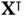 , but the theoretical complexity is only a part of the story. Matrix–vector multiplications are amenable to a variety of hardware optimizations, which in practice can make orders of magnitude difference in speed ( Section 4.2). In fact, given how arduous manually optimizing computational bottlenecks can be, designing algorithms so as to take advantage of common routines (as those in Level 3 BLAS) and their ready‐optimized implementations has been recognized as an effective principle in algorithm design [65].
, but the theoretical complexity is only a part of the story. Matrix–vector multiplications are amenable to a variety of hardware optimizations, which in practice can make orders of magnitude difference in speed ( Section 4.2). In fact, given how arduous manually optimizing computational bottlenecks can be, designing algorithms so as to take advantage of common routines (as those in Level 3 BLAS) and their ready‐optimized implementations has been recognized as an effective principle in algorithm design [65].
3.2 Phylogenetic Reconstruction
While big 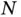 and big
and big 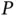 regression adapts a classical statistical task to contemporary needs, the twenty‐first century is witnessing the application of computational statistics to the entirety of applied science. One such example is the tracking and reconstruction of deadly global viral pandemics. Molecular phylogenetics has become an essential analytical tool for understanding the complex patterns in which rapidly evolving pathogens propagate throughout and between countries, owing to the complex travel and transportation patterns evinced by modern economies [66], along with other factors such as increased global population and urbanization [67]. The advance in sequencing technology is generating pathogen genomic data at an ever‐increasing pace, with a trend to real time that requires the development of computational statistical methods that are able to process the sequences in a timely manner and produce interpretable results to inform national/global public health organizations.
regression adapts a classical statistical task to contemporary needs, the twenty‐first century is witnessing the application of computational statistics to the entirety of applied science. One such example is the tracking and reconstruction of deadly global viral pandemics. Molecular phylogenetics has become an essential analytical tool for understanding the complex patterns in which rapidly evolving pathogens propagate throughout and between countries, owing to the complex travel and transportation patterns evinced by modern economies [66], along with other factors such as increased global population and urbanization [67]. The advance in sequencing technology is generating pathogen genomic data at an ever‐increasing pace, with a trend to real time that requires the development of computational statistical methods that are able to process the sequences in a timely manner and produce interpretable results to inform national/global public health organizations.
The previous three Core Challenges are usually interwound such that the increase in the sample size (big N) and the number of traits (big P) for each sample usually happen simultaneously and lead to increased heterogeneity that requires more complex models (big M). For example, recent studies in viral evolution have seen a continuing increase in the sample size that the West Nile virus, Dengue, HIV, and Ebola virus studies involve 104, 352, 465, and 1610 sequences [68–71], and the GISAID database has collected 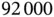 COVID‐19 genomic sequences by the end of August 2020 [72].
COVID‐19 genomic sequences by the end of August 2020 [72].
To accommodate the increasing size and heterogeneity in the data and be able to apply the aforementioned efficient gradient‐based algorithms, Ji et al . [73] propose a linear‐time algorithm for calculating an 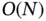 ‐dimensional gradient on a tree w.r.t. the sequence evolution. The linear‐time gradient algorithm calculates each branch‐specific derivative through a preorder traversal that complements the postorder traversal from the likelihood calculation of the observed sequence data at the tip of the phylogeny by marginalizing over all possible hidden states on the internal nodes. The pre‐ and postorder traversals complete the Baum's forward–backward algorithm in a phylogenetic framework [74]. The authors then apply the gradient algorithm with HMC ( Section 2.2) samplers to learn the branch‐specific viral evolutionary rates.
‐dimensional gradient on a tree w.r.t. the sequence evolution. The linear‐time gradient algorithm calculates each branch‐specific derivative through a preorder traversal that complements the postorder traversal from the likelihood calculation of the observed sequence data at the tip of the phylogeny by marginalizing over all possible hidden states on the internal nodes. The pre‐ and postorder traversals complete the Baum's forward–backward algorithm in a phylogenetic framework [74]. The authors then apply the gradient algorithm with HMC ( Section 2.2) samplers to learn the branch‐specific viral evolutionary rates.
Читать дальше

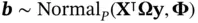 by first generating
by first generating 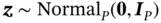 and
and 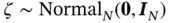 and then setting
and then setting 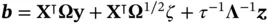 and (ii) subsequently solving
and (ii) subsequently solving 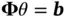 yields a sample
yields a sample 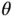 from the distribution ( 4). The authors then observe that the mechanism through which a shrinkage prior induces sparsity of
from the distribution ( 4). The authors then observe that the mechanism through which a shrinkage prior induces sparsity of 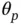 s also induces a tight clustering of eigenvalues in the prior‐preconditioned matrix
s also induces a tight clustering of eigenvalues in the prior‐preconditioned matrix 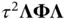 . This fact makes it possible for prior‐preconditioned CG to solve the system
. This fact makes it possible for prior‐preconditioned CG to solve the system  in
in 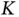 matrix–vector operations of form
matrix–vector operations of form 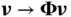 , where
, where 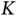 roughly represents the number of significant
roughly represents the number of significant 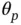 s that are distinguishable from zeros under the posterior. For
s that are distinguishable from zeros under the posterior. For 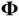 having a structure as in ( 4),
having a structure as in ( 4),  can be computed via matrix–vector multiplications of form
can be computed via matrix–vector multiplications of form 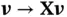 and
and 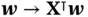 , so each
, so each 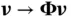 operation requires a fraction of the computational cost of directly computing
operation requires a fraction of the computational cost of directly computing  and then factorizing it.
and then factorizing it.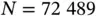 patients and
patients and 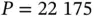 covariates [57]. Though unexplored in their work, the algorithm's heavy use of matrix–vector multiplications provides avenues for further acceleration. Technically, the algorithm's complexity may be characterized as
covariates [57]. Though unexplored in their work, the algorithm's heavy use of matrix–vector multiplications provides avenues for further acceleration. Technically, the algorithm's complexity may be characterized as 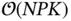 , for the
, for the 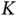 matrix–vector multiplications by
matrix–vector multiplications by 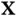 and
and 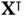 , but the theoretical complexity is only a part of the story. Matrix–vector multiplications are amenable to a variety of hardware optimizations, which in practice can make orders of magnitude difference in speed ( Section 4.2). In fact, given how arduous manually optimizing computational bottlenecks can be, designing algorithms so as to take advantage of common routines (as those in Level 3 BLAS) and their ready‐optimized implementations has been recognized as an effective principle in algorithm design [65].
, but the theoretical complexity is only a part of the story. Matrix–vector multiplications are amenable to a variety of hardware optimizations, which in practice can make orders of magnitude difference in speed ( Section 4.2). In fact, given how arduous manually optimizing computational bottlenecks can be, designing algorithms so as to take advantage of common routines (as those in Level 3 BLAS) and their ready‐optimized implementations has been recognized as an effective principle in algorithm design [65].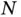 and big
and big 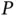 regression adapts a classical statistical task to contemporary needs, the twenty‐first century is witnessing the application of computational statistics to the entirety of applied science. One such example is the tracking and reconstruction of deadly global viral pandemics. Molecular phylogenetics has become an essential analytical tool for understanding the complex patterns in which rapidly evolving pathogens propagate throughout and between countries, owing to the complex travel and transportation patterns evinced by modern economies [66], along with other factors such as increased global population and urbanization [67]. The advance in sequencing technology is generating pathogen genomic data at an ever‐increasing pace, with a trend to real time that requires the development of computational statistical methods that are able to process the sequences in a timely manner and produce interpretable results to inform national/global public health organizations.
regression adapts a classical statistical task to contemporary needs, the twenty‐first century is witnessing the application of computational statistics to the entirety of applied science. One such example is the tracking and reconstruction of deadly global viral pandemics. Molecular phylogenetics has become an essential analytical tool for understanding the complex patterns in which rapidly evolving pathogens propagate throughout and between countries, owing to the complex travel and transportation patterns evinced by modern economies [66], along with other factors such as increased global population and urbanization [67]. The advance in sequencing technology is generating pathogen genomic data at an ever‐increasing pace, with a trend to real time that requires the development of computational statistical methods that are able to process the sequences in a timely manner and produce interpretable results to inform national/global public health organizations.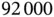 COVID‐19 genomic sequences by the end of August 2020 [72].
COVID‐19 genomic sequences by the end of August 2020 [72].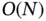 ‐dimensional gradient on a tree w.r.t. the sequence evolution. The linear‐time gradient algorithm calculates each branch‐specific derivative through a preorder traversal that complements the postorder traversal from the likelihood calculation of the observed sequence data at the tip of the phylogeny by marginalizing over all possible hidden states on the internal nodes. The pre‐ and postorder traversals complete the Baum's forward–backward algorithm in a phylogenetic framework [74]. The authors then apply the gradient algorithm with HMC ( Section 2.2) samplers to learn the branch‐specific viral evolutionary rates.
‐dimensional gradient on a tree w.r.t. the sequence evolution. The linear‐time gradient algorithm calculates each branch‐specific derivative through a preorder traversal that complements the postorder traversal from the likelihood calculation of the observed sequence data at the tip of the phylogeny by marginalizing over all possible hidden states on the internal nodes. The pre‐ and postorder traversals complete the Baum's forward–backward algorithm in a phylogenetic framework [74]. The authors then apply the gradient algorithm with HMC ( Section 2.2) samplers to learn the branch‐specific viral evolutionary rates.![Роман Зыков - Роман с Data Science. Как монетизировать большие данные [litres]](/books/438007/roman-zykov-roman-s-data-science-kak-monetizirova-thumb.webp)










