Despite the previously mentioned limitations, profiling of microbiotas based on shorter variable regions of the 16S rRNA gene using current sequencing technologies has proven invaluable in providing insights into the role of microbiota in health and disease and how our microbiotas change over time and under different conditions. For instance, such profiling analysis using species-level classification has been used to compare the vaginal microbiota of women of African-American versus European ancestry ( Figure 5-9). The results revealed significant differences in the microbiota composition of these two groups of women. The profiles confirmed that women of European ancestry are more likely to have a Lactobacillus-dominated vaginal microbiota, whereas African-American women are more likely to have a more diverse vaginal microbiota, similar to a composition often found in women displaying symptoms of BV. Even at the species level, significant differences were observed within the Lactobacillus genus, with women of European ancestry more likely to harbor L. crispatus, L. jensenii, and L. gasseri. These findings supported other studies indicating that African-American women are more prone to having diseases that are associated with BV, such as increased risk of sexually transmitted diseases and adverse pregnancy outcomes. Researchers are now using this information to understand the factors that dictate these differences and how to develop treatment options that take these differences into account.
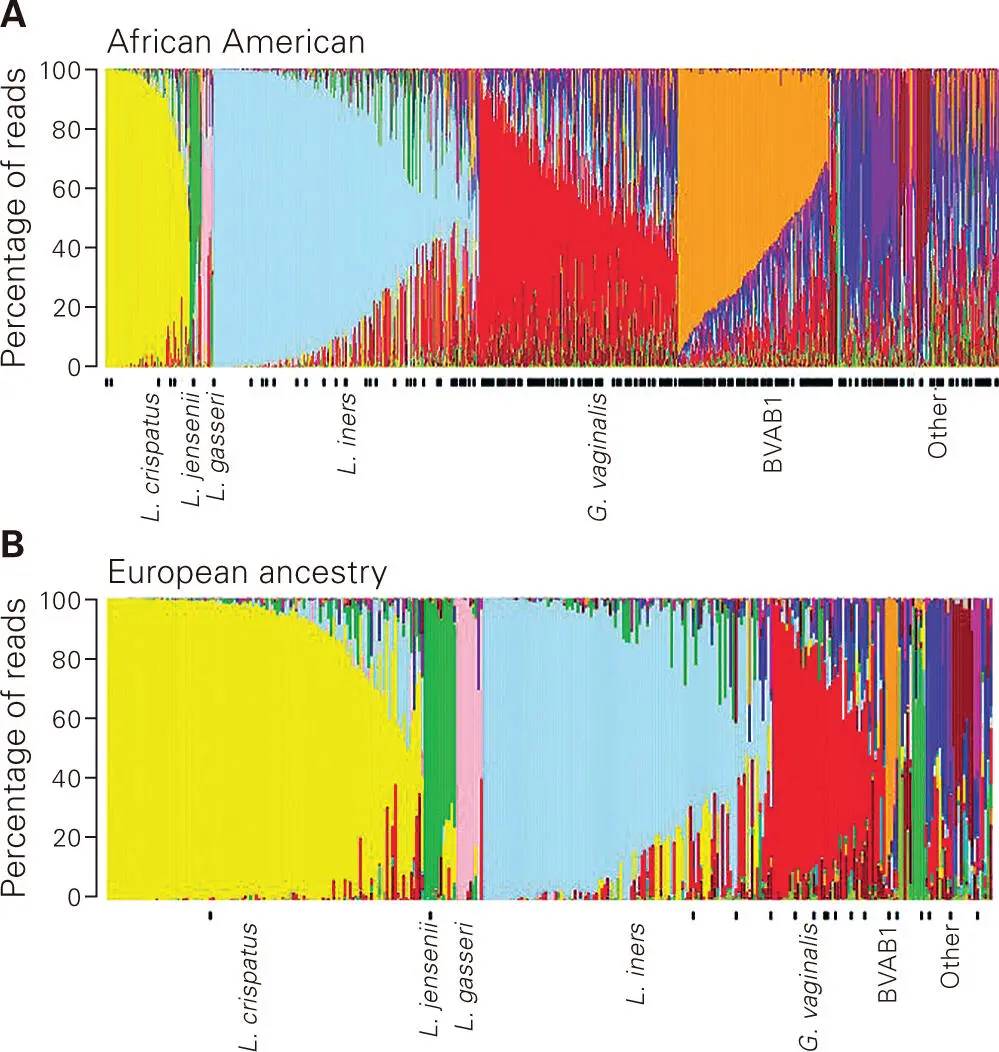
Figure 5-9. Comparative 16S rRNA gene-based profiling of bacterial communities from women of African-American versus European ancestry. Relative abundance profiles of vaginal bacteria using species-level classification from (A) 960 African-American women and (B) 330 women of European ancestry enrolled in the Vaginal Human Microbiome Project. Each vertical line represents the bacterial composition profile from one vaginal sample, where the profiles are grouped by the dominant species into different profile types. The colors correlate to different bacterial species: yellow = Lactobacillus crispatus; green = Lactobacillus jensenii; pink = Lactobacillus gasseri; light blue = Lactobacillus iners; red = Gardnerella vaginalis; orange = uncultivated clostridial species associated with bacterial vaginosis type 1 (BVAB1); dark blue = Prevotella species. The black bars underneath the plot indicate microbial profiles from women diagnosed with bacterial vaginosis (BV). Reprinted from Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, Strauss JF 3rd; Vaginal Microbiome Consortium, Jefferson KK, Buck GA. 2014. Microbiology 160(pt10):2272–2282, with permission.
Tracking Microbes Through Space and Time by Multilocus Sequence Typing (MLST) and Whole-Genome-Sequencing. We have considered the problem of identifying different bacterial species in complex mixtures taken from the environment or from sites in the human body. But scientists and epidemiologists are often faced with the problem of distinguishing different isolates or variants of a single bacterial species. Suppose that there is an outbreak of Staphylococcus aureus in a hospital, and you need to trace how this particular strain got into and around the hospital. Was it carried by a hospital worker or by a family member of a patient? Or suppose there is an outbreak of food poisoning caused by Listeria monocytogenes, which can contaminate food-processing equipment and ready-to-eat meat products. You want to find out how this strain of L. monocytogenes entered the food chain and whether it is similar to strains that caused previous outbreaks. The rRNA genes of all isolates of S. aureus have the same sequence, because the rRNA genes change very slowly within a given species. Likewise, the DNA sequences of the rRNA genes are nearly identical for different variants and isolates of L. monocytogenes.
Clearly, 16S rRNA gene-based strain typing will not work to trace infections such as these. But unlike rRNA genes, the DNA sequences of genes that encode housekeeping enzymes and virulence factors do change with time within a species. That is, isolates from S. aureus or L. monocytogenes from different locations or sources accumulate slight differences in the sequences in their housekeeping and virulence factor genes over time. This drift arises partly because the genetic code is degenerate. Recall that more than one codon can specify the same amino acid (e.g., there are six specifying leucine), and much of this redundancy occurs at the third positions of codons. Therefore, the DNA sequences of housekeeping genes can show variations in different isolates of a given bacterial species and still specify the same amino acid in the enzyme product. The drift in virulence factor genes often involves changes in amino acids compared to those in housekeeping genes, because the virulence factor genes are subjected to strong selective pressures during infection.
This variation within coding sequences is the basis of multilocus sequence typing (MLST), but rather than relying on genetic drift in just one housekeeping or virulence gene, the DNA sequences of regions in multiple (usually seven or more) genes are analyzed for each isolate. For each of the genes selected for a particular bacterial species, the different sequences are assigned as alleles. MLST analysis using multiple genes is easy and has become cost-effective recently. Bioinformatics is used to identify regions of seven or more housekeeping or virulence genes that show variations in a given bacterial species. Pairs of PCR primers are designed to amplify and sequence about 500 base pairs from each of these variable regions. The sequences of these multiple loci can then be compared among samples or with other isolates in databases to trace the relatedness of the different isolates locally and worldwide. This analysis can also take into account combinations of alleles in bacterial species that exchange genetic material and undergo horizontal gene transfer frequently.
Going back to our examples, samples collected from staff, visitors, patients, and locations in the hospital could be cultured to identify those that contain S. aureus, which is a common commensal bacterium that is easily identified on growth medium. The different isolates of S. aureus would be subjected to MLST analysis. Progeny or clonal isolates will have the same DNA sequences in most of the multiple loci, whereas strains from a different source may have loci with sequence variations. The resulting profile would indicate whether patients are infected with the same strain of S. aureus and where this strain may have arisen in the hospital. Similarly, MLST analysis can trace the sources of L. monocytogenes through the chain of food preparation of the outbreak mentioned earlier as well as previous outbreaks and thereby identify the root of the contamination problem.
MLST analysis does not distinguish whether multiple sequence differences observed between alleles is a result of multiple point mutations or a single recombination event. Consequently, MLST analysis does not assign a higher similarity value to sequences differing by a single nucleotide compared to sequences with multiple nucleotide differences. To determine phylogenetic relationships of closely related species with high clonal evolution, multilocus sequence analysis (MLSA) is used, in which the selected housekeeping and virulence gene sequences are first concatenated (i.e., virtually linked in tandem to each other) before performing comparative phylogenetic analysis. This process provides greater discriminatory power for determining phylogenetic relationships.
MLSA analysis has recently been greatly extended by using whole-genome sequence determinations, rather than sequences of a limited number of housekeeping or virulence genes. MLSA by whole-genome sequencing provides higher resolution for differentiating bacterial isolates and for tracking the evolution and spread of virulent and/or antibiotic-resistant bacteria. As an example of the impact that whole-genome sequencing has had on MLSA, let us consider carbapenem-resistant Klebsiella pneumoniae, which has emerged as a serious clinical problem in hospitals. Previously, the vast majority of resistant clinical isolates had been genetically characterized by MLST as a single multilocus type (designated as ST258), leading to the hypothesis that a single clone of ST258 was responsible for the global spread of these multidrug-resistant bacteria. However, whole-genome sequencing and phylogenetic analysis of a large number of ST258 clinical isolates from diverse geographic locations revealed unexpected diversity among the isolates, including divergence into two distinct genetic clades due to a 215-kB region that appears to be a hotspot for DNA recombination events. This region also contains genes involved in capsule polysaccharide biosynthesis that likely contributed to the observed serological variation. In another example, phylogenetic relationship analysis based on whole-genome sequencing of 132 clinical isolates from around the world was used to track the recent evolutionary history of the dysentery (bloody diarrhea) pathogen Shigella sonnei from Europe to other parts of the world ( Figure 5-10).
Читать дальше













