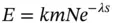As the extension continues, at some point, mismatches and gaps will begin to outweigh the exact matches and conservative substitutions, accruing negative scores from the scoring matrix. As soon as the curve begins to turn downward, BLAST measures whether the drop-off exceeds a threshold called X . If the curve decays more than is allowed by the value of X , the extension is terminated and the alignment is trimmed back to the length corresponding to the preceding maximum in the curve. The resulting alignment is called a high-scoring segment pair , or HSP. Given that the BLAST algorithm systematically marches across the query sequence using all possible query words, it is possible that more than one HSP may be found for any given sequence pair.
After an HSP is identified, it is important to determine whether the resulting alignment is actually significant. Using the cumulative score from the alignment, along with a number of other parameters, a new value called E (for “expect”) is calculated ( Box 3.2). For each hit, E gives the number of expected HSPs having a score of S or more that BLAST would find purely by chance. Put another way, the value of E provides a measure of whether the reported HSP is a false positive (see Box 5.4). Lower E values imply greater biological significance.
Box 3.2The Karlin–Altschul Equation
As one might imagine, assessing the putative biological significance of any given BLAST hit based simply on raw scores is difficult, since the scores are dependent on the composition of the query and target sequences, the length of the sequences, the scoring matrix used to compute the raw scores, and numerous other factors. In one of the most important papers on the theory of local sequence alignment statistics, Karlin and Altschul (1990) presented a formula which directly addresses this problem. The formula, which has come to be known as the Karlin–Altschul equation, uses search-specific parameters to calculate an expectation value ( E ). This value represents the number of HSPs that would be expected purely by chance. The equation and the parameters used to calculate E are as follows:
where k is a minor constant, m is the number of letters in the query, N is the total number of letters in the target database, λ is a constant used to normalize the raw score of the high-scoring segment pair, with the value of λ varying depending on the scoring matrix used; and S is the score of the high-scoring segment pair.
Performing a BLAST Search
While many BLAST servers are available throughout the world, the most widely used portal for these searches is the BLAST home page at the National Center for Biotechnology Information (NCBI; Figure 3.5). The top part of the page provides access to the most frequently performed types of BLAST searches, summarized in Table 3.2, while the lower part of the page is devoted to specialized types of BLAST searches. To illustrate the relative ease with which one can perform a BLAST search, a protein-based search using BLASTP is discussed. Clicking on the Protein BLAST box brings users to the BLASTP search page, a portion of which is shown in Figure 3.6. Obviously, a query sequence that will be used as the basis for comparison is required. Harking back to the Entrez discussion in Chapter 2, the sequence of the netrin receptor from Homo sapiens (NP_005206.2) has been pasted into the query sequence box. Immediately to the right, the user can use the query subrange boxes to specify whether only a portion of this sequence is to be used; if the whole sequence is to be used, these fields should be left blank.
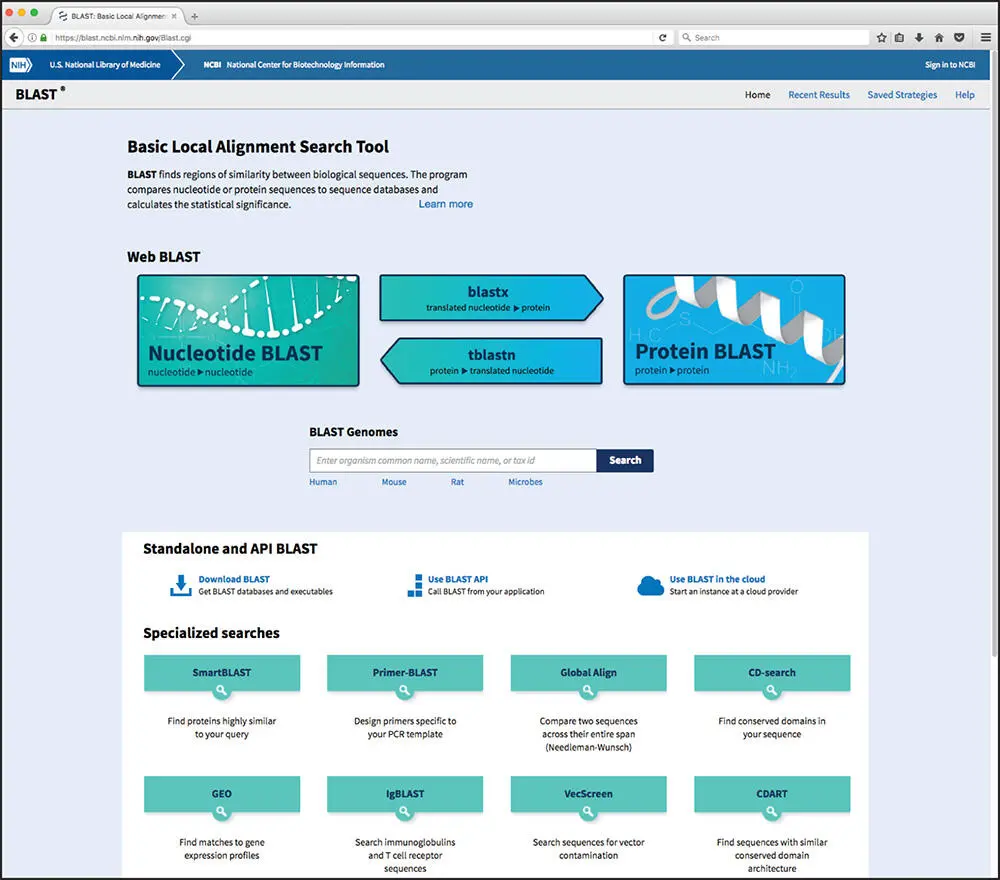
Figure 3.5 The National Center for Biotechnology Information (NCBI) BLAST landing page. Examples of the most commonly used queries that can be performed using the BLAST interface are discussed in the text.
Moving to the Choose Search Set section of the page, the database to be searched can be selected using the Database pull-down menu; clicking on the question mark next to the Database pull-down provides a brief description of each of the available target databases. Here, the search will be performed against the RefSeq database (see Box 1.2). Directly below, the Organism box can be used to limit the search results to sequences from individual organisms or taxa. While not part of this worked example, if the user wanted to limit the returned results to those from just mouse and rat, using the same type of syntax used in issuing Entrez searches (see Table 2.1), the user would type Mus musculus [ORGN] AND Rattus norvegicus [ORGN]in this field; if the user wanted all results except those from mouse and rat, they would also need to check the Exclude box. As this search will be performed against RefSeq, one can exclude predicted proteins from the search results by clicking the “Models (XM/XP)” checkbox. Finally, in the Program Selection section, BLASTP is selected by default.
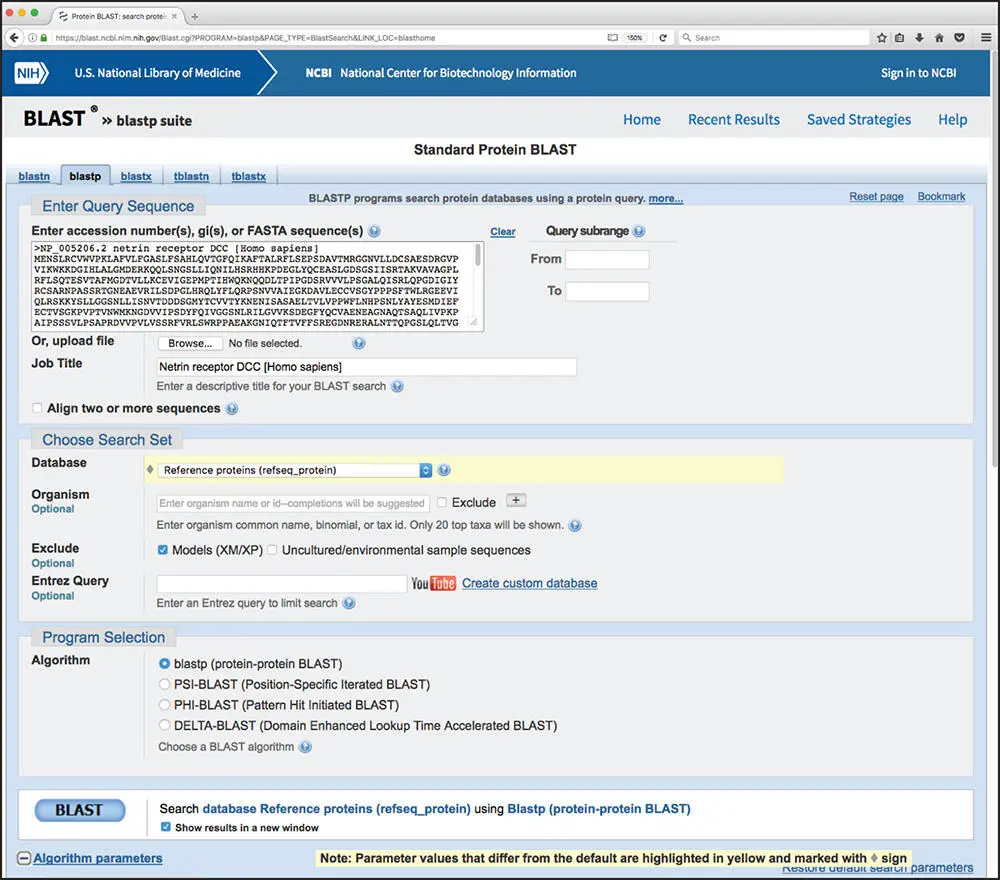
Figure 3.6 The upper portion of the BLASTP query page. The first section in the window is used to specify the sequence of interest, whether only a portion of that sequence should be used in performing the search (query subrange), which database should be searched, and which protein-based BLAST algorithm should be used to execute the query. See text for details.
If the user wishes to use the default settings for all algorithm parameters, the search can be submitted by simply clicking on the blue BLAST button. However, the user can exert finer control over how the search is performed by changing the items found in the Algorithm parameters section. To access these settings, the user must first click on the plus sign next to the words “Algorithm parameters” to expand this section of the web page, producing the view shown in Figure 3.7. This part of the query page is where the theory underlying a BLAST search discussed earlier in this chapter comes into play. In the General Parameters section, the expect threshold limits returned results to those having an E value lower than the specified value, with smaller values providing a more stringent cut-off. The word size setting changes the size of the query word used to initiate the BLAST search, with longer word sizes initiating the search with longer ungapped alignments. A word size of 3 is recommended for protein searches, as shorter words increase sensitivity; however, if searching for near-exact matches, a longer word size can be used, also yielding faster search times.

Figure 3.7 The lower portion of the BLASTP query page, showing algorithm parameters that the user can adjust to fine-tune the search. Values that have been changed for the search discussed in the text are highlighted in yellow and marked with a diamond. See text for details.
In the Scoring Parameters section, the user can select an appropriate scoring matrix (with the default being BLOSUM62). Changing the matrix automatically changes the gap penalties to values appropriate for that scoring matrix. As described in the discussion of affine gap penalties above, the user may change these values manually; increasing the gap costs would result in pairwise alignments with fewer gaps, where decreasing the values would make the insertion of gaps more permissive.
Читать дальше