The scoring of gaps in pairwise sequence alignments is different from scoring approaches discussed to this point, as no comparison between characters is possible – one sequence has a residue at some position and the other sequence has nothing. The most widely used method for scoring gaps involves a quantity known as the affine gap penalty . Here, a fixed deduction is made for introducing the gap; an additional deduction is made that is proportional to the length of the gap. The formula for the affine gap penalty is G + Ln , where G is the gap-opening penalty (the cost of creating the gap), L is the gap-extension penalty, and n is the length of the gap, with G > L . This last condition is important: given that the gap-opening penalty is larger than the gap-extension penalty, lengthening existing gaps would be favored over creating new ones. The values of G and L can be adjusted manually in most programs to make the insertion of gaps either more or less permissive, but most methods automatically adjust both G and L to the most appropriate values for the scoring matrix being used.

Figure 3.2 A nucleotide scoring table. The scoring for the four nucleotide bases is shown in the upper left of the figure, with the remaining one-letter codes specifying the IUPAC/UBMB codes for ambiguities or chemical similarities. Note that the matrix is a mirror image of itself with respect to the diagonal.
The other major type of gap penalty used is a non-affine (or linear ) gap penalty . Here, there is no cost for opening the gap; a simple, fixed mismatch penalty is assessed for each position in the gap. It is thought that affine penalties better represent the biology underlying the sequence alignments, as affine gap penalties take into account the fact that most conserved regions are ungapped and that a single mutational event could insert or delete many more than just one residue. In practice, use of the affine gap penalty better enables the detection of more distant homologs.
By far the most widely used technique for detecting similarity between sequences of interest is the Basic Local Alignment Search Tool, or BLAST (Altschul et al. 1991). The widespread adoption of BLAST as a cornerstone technique in sequence analysis lies in its ability to detect similarities between nucleotide and protein sequences accurately and quickly, without sacrificing sensitivity. The original, standard family of BLAST programs is shown in Table 3.2, but in the time since its introduction many variations of the original BLAST program have been developed to address specific needs in the realm of pairwise sequence comparison, several of which will be discussed in this chapter.
BLAST is a local alignment method that is capable of detecting not only the best region of local alignment between a query sequence and its target, but also whether there are other plausible alignments between the query and the target. To find these regions of local alignment in a computationally efficient fashion, the method begins by seeding the search with a small subset of letters from the query sequence, known as the query word . Using the example shown in Figure 3.3, consider a search where the query word of default length 3 is RDQ. (In practice, all words of length 3 are considered, so, using the sequence in Figure 3.3, the first query word would be TLS, followed by LSH, and so on across the sequence.) BLAST now needs to find not only the word RDQ in all of the sequences in the target database but also related words where conservative substitutions have been introduced, as those matches may also be biologically informative and relevant. To determine which words are related to RDQ, scoring matrices are used to develop what is called the neighborhood. The center panel of Figure 3.3shows the collection of words that are related to the original query word, in descending score order; the scores here are calculated using a BLOSUM62 scoring matrix ( Figure 3.1). Obviously, some cut-off must be applied so that further consideration is only given to words that are indeed closely related to the original query word. The parameter that controls this cut-off is the neighborhood score threshold ( T ). The value of T is determined automatically by the BLAST program but can be adjusted by the user. Increasing T would push the search toward more exact matches and would speed up the search, but could lead to overlooking possibly interesting biological relationships. Decreasing T allows for the detection of more distant relationships between sequences. Here, only words with T ≥ 11 move to the next step.
Table 3.2 BLAST algorithms.
| Program |
Query |
Database |
| BLASTN |
Nucleotide |
Nucleotide |
| BLASTP |
Protein |
Protein |
| BLASTX |
Nucleotide, six-frame translation |
Protein |
| TBLASTN |
Protein |
Nucleotide, six-frame translation |
| TBLASTX |
Nucleotide, six-frame translation |
Nucleotide, six-frame translation |

Figure 3.3 The initiation of a BLAST search. The search begins with query words of a given length (here, three amino acids) being compared against a scoring matrix to determine additional three-letter words “in the neighborhood” of the original query word. Any occurrences of these neighborhood words in sequences within the target database are then investigated. See text for details.
Focusing now on the lower panel of Figure 3.3, the original query word (RDQ) has been aligned with another word from the neighborhood whose score is more than the score threshold of T ≥ 11 (REQ). The BLAST algorithm now attempts to extend this alignment in both directions, tallying a cumulative score resulting from matches, mismatches, and gaps, until it constructs a local alignment of maximal length. Determining what the maximal length actually is can be best explained by considering the graph in Figure 3.4. Here, the number of residues that have been aligned is plotted against the cumulative score resulting from the alignment. The left-most point on the graph represents the alignment of the original query word with one of the words from the neighborhood, again having a value of T = 11 or greater. As the extension proceeds, as long as exact matches and conservative substitutions outweigh mismatches and gaps, the cumulative score will increase. As soon as the cumulative score breaks the score threshold S , the alignment is reported in the BLAST output. Simply clearing S does not automatically mean that the alignment is biologically significant, a very important point that will be addressed later in this discussion.
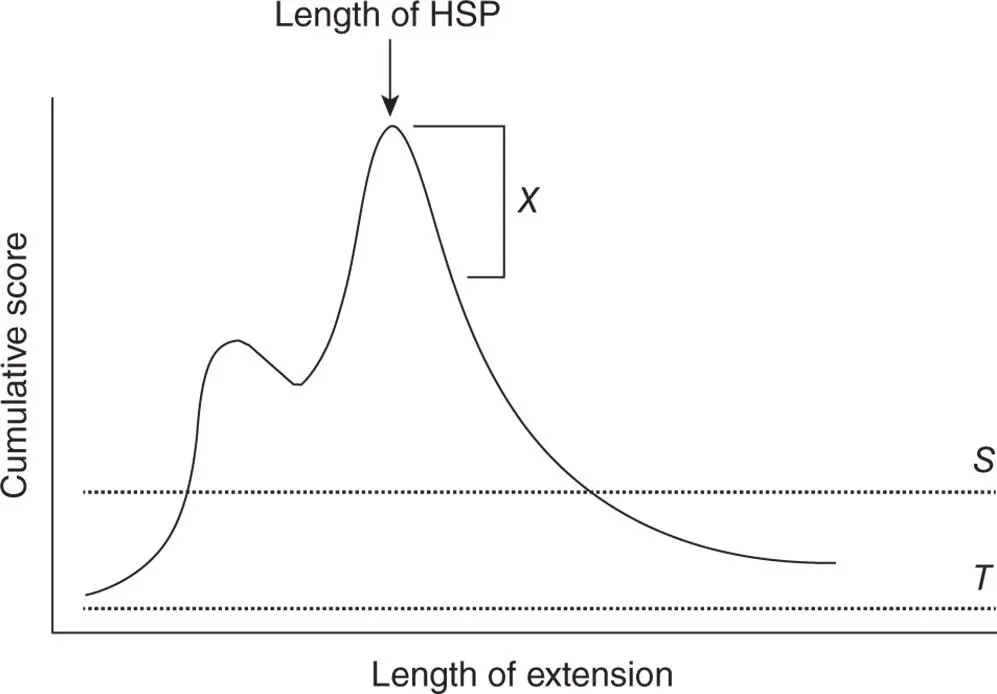
Figure 3.4 BLAST search extension. Length of extension represents the number of characters that have been aligned in a pairwise sequence comparison. Cumulative score represents the sum of the position-by-position scores, as determined by the scoring matrix used for the search. T represents the neighborhood score threshold, S is the minimum score required to return a hit in the BLAST output, and X is the significance decay. See text for details.
Читать дальше







