Jane Flint - Principles of Virology
Здесь есть возможность читать онлайн «Jane Flint - Principles of Virology» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Principles of Virology
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:3 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 60
- 1
- 2
- 3
- 4
- 5
Principles of Virology: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Principles of Virology»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
Volume I: Molecular Biology
Volume II: Pathogenesis and Control
Principles of Virology, Fifth Edition
Principles of Virology — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Principles of Virology», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
Other Envelope Proteins
The envelopes of some viruses, including orthomyxoviruses, herpesviruses, and poxviruses, contain integral membrane proteins that lack large external domains or possess multiple membrane-spanning segments. Among the best characterized is the influenza A virus M2 protein. This small (97-amino-acid) protein is a minor component of virus particles. In the viral membrane, two disulfide-linked M2 dimers associate to form a noncovalent tetramer that functions as an ion channel. This viral ion channel is the target of the influenza virus inhibitor drug amantadine (Volume II, Fig. 9.13). The effects of this drug, as well as of mutations in the M2 coding sequence, indicate that M2 plays important roles during both entry, by controlling the pH of the virus particle interior ( Chapter 5), and release of newly assembled virus particles ( Chapter 13). M2 belongs to a class of channel-creating viral proteins called viroporins, which are present in a number of other enveloped viruses, such as hepatitis C virus and Sindbis virus, but also in nonenveloped viruses like simian virus 40 and papillomaviruses.
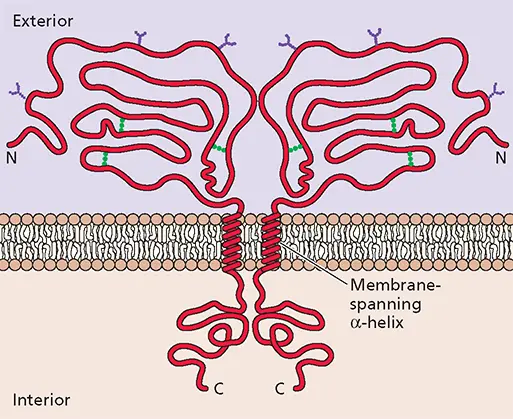
Figure 4.22 Structural and chemical features of a typical viral envelope glycoprotein shown schematically. The protein is inserted into the lipid bilayer via a single membrane-spanning domain. This segment separates a larger external domain, which is decorated with N-linked oligosaccharides (purple) and contains disulfide bonds (green), from a smaller internal domain.
Simple Enveloped Viruses: Direct Contact of External Proteins with the Capsid or Nucleocapsid
In the simplest enveloped viruses, exemplified by (+) strand RNA alphaviruses such as Semliki Forest, Sindbis, and Ross River viruses, the envelope directly abuts an inner nucleocapsid containing the (+) strand RNA genome. This inner protein layer is a T = 4 icosahedral shell built from 240 copies of a single capsid (C) protein arranged as hexamers and pentamers. The outer layer of the envelope also contains 240 copies of the viral glycoproteins E1 and E2, which form heterodimers. These heterodimers cover the surface of the particle, such that the lipid membrane is not exposed on the exterior. Strikingly, the glycoproteins are also organized into a T = 4 icosahedral shell ( Fig. 4.24A).
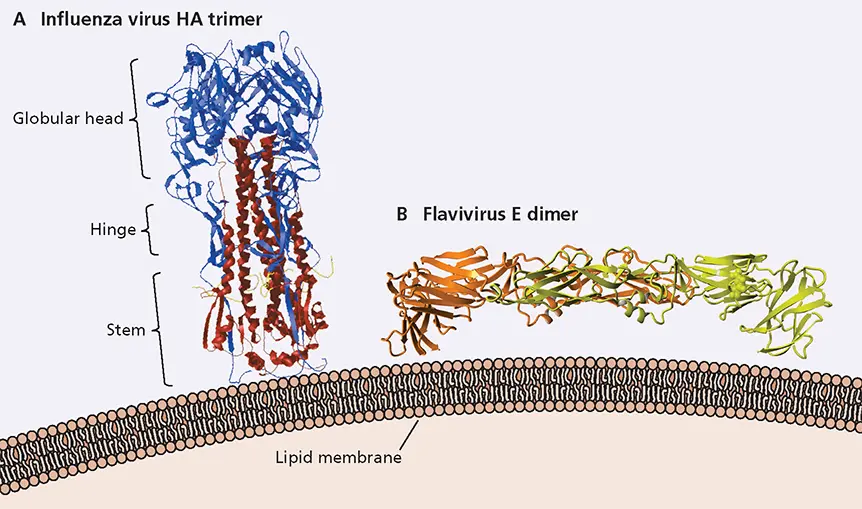
Figure 4.23 Structures of extracellular domains of viral glycoproteins. These extracellular domains, which were cleaved from transmembrane and internal domain for crystallization, are depicted as they are oriented with respect to the membrane of the viral envelope. (A)X-ray crystal structure of the influenza virus HA glycoprotein trimer. Each monomer comprises HA1 (blue) and HA2 (red) subunits covalently linked by a disulfide bond. Data from Chen J et al. 1998. Cell 95:409–417, with permission. (B)X-ray structure of the tick-borne encephalitis virus (a flavivirus) E protein dimer, with the subunits shown in orange and yellow. PDB ID: 1SVB. Data from Rey FA, Harrison SC. 1995. Nature 375:291–298.
The structure of Sindbis virus has been determined by cryo-EM and image reconstruction to some 9-Å resolution ( Fig. 4.24A), while the structures of the E1 and C proteins of the related Semliki Forest virus have been solved at high resolution. The organization of the alphavirus envelope, including the transmembrane anchoring of the outer glycoprotein layer to structural units of the nucleocapsid, can therefore be described with unprecedented precision. The transmembrane segments of the E1 and E2 glycoproteins form a pair of tightly associated α-helices, with the cytoplasmic domain of E2 in close apposition to a cleft in the capsid protein ( Fig. 4.24Band C). This interaction accounts for the 1:1 symmetry match between the internal capsid and exterior glycoproteins. On the outer surface of the membrane, the external portions of these glycoproteins, together with the E3 protein, form an unexpectedly elaborate structure: a thin T = 4 icosahedral protein layer covers most of the membrane ( Fig. 4.24Aand B) and supports the spikes, which are hollow, three-lobed projections ( Fig. 4.24C).
The structures formed by external domains of membrane proteins of the important human pathogens West Nile virus and dengue virus (family Flaviviridae ) are quite different: they lie flat on the particle surface rather than forming protruding spikes ( Fig. 4.25; see also Box 4.9). Nevertheless, the alphavirus E1 protein and the single flavivirus envelope (E) protein exhibit the same topology ( Fig. 4.25A), suggesting that the genes encoding them evolved from a common ancestor. Furthermore, the external domains of flaviviral E proteins are also icosahedrally ordered, and the envelopes of viruses of these families are described as structured. In contrast, as described in the next section, the arrangement of membrane proteins generally exhibits little relationship to the structure of the capsid or internal nucleoprotein when virus particles contain additional protein layers.
Enveloped Viruses with an Additional Protein Layer
Enveloped viruses of several families contain an additional protein layer that mediates interactions of the genome-containing structure with the viral envelope. In the simplest case, a single viral protein, termed the matrix protein, welds an internal ribonucleoprotein to the envelope. This arrangement is found in members of several groups of (−) strand RNA viruses ( Fig. 4.6C; Appendix, Fig. 17 and 31). Retrovirus particles also contain an analogous, membrane-associated matrix protein (MA), which is closely associated with the inner surface of the viral envelope.
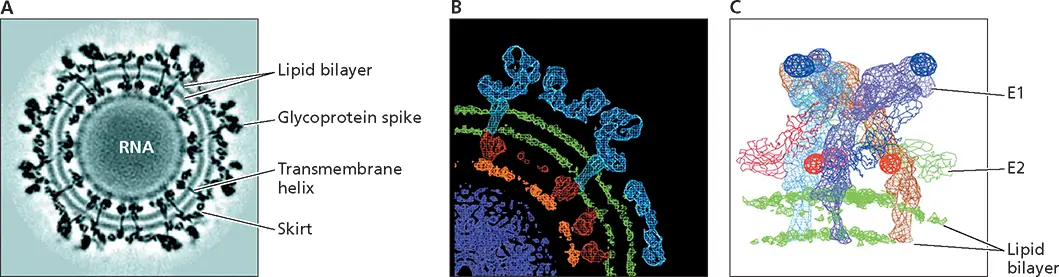
Figure 4.24 Structure of a simple enveloped virus, Sindbis virus. (A)Cross section through the cryo-EM density map of Sindbis virus, a member of the alphavirus genus of the Togaviridae , at 11-Å resolution. The lipid bilayer of the viral envelope is clearly defined at this resolution, as are the transmembrane domains of the glycoproteins. (B)Different layers of the particle, based on the fitting of a high-resolution structure of the E1 glycoprotein into a 9-Å reconstruction of the virus particle. The nucleocapsid (red) (C protein) surrounds the genomic (+) strand RNA. The RNA is the least well-ordered feature in the reconstruction, although segments (orange) lying just below the capsid protein appear to be ordered by interaction with this protein. The C protein penetrates the inner leaflet of the lipid membrane, where it interacts with the cytoplasmic domain of the E2 glycoprotein (blue). The membrane is spanned by rod-like structures that are connected to the skirt (see panel A) by short stems. (C)The structure of the E1 and E2 glycoproteins, obtained by fitting the crystal structure of the closely related Semliki Forest virus E1 glycoprotein into the 11-Å density map and assigning density unaccounted for to the E2 glycoprotein. The three E2 glycoprotein molecules in a trimeric spike are colored light blue, dark blue, and brown, and the E1 molecules shown as backbone traces colored red, green, and magenta. The portions of the proteins that cross the lipid bilayer are helical. Adapted from Zhang W et al. 2002. J Virol 76:11645–11658, with permission. Courtesy of Michael Rossmann, Purdue University.
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Principles of Virology»
Представляем Вашему вниманию похожие книги на «Principles of Virology» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Principles of Virology» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.











