1 Ciccarelli, F.D. (2006). Toward automatic reconstruction of a highly resolved tree of life. Science 311 (5765): 1283–1287.
2 Letunic, I. (2007). Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23 (1): 127–128.
1 Alberts, B., Johnson, A., Lewis, L. et al. (2015). Molecular Biology of the Cell, 6e. New York: Garland Science.
2 Alberts, B., Bray, D., Hopkin, K. et al. (2019). Essential Cell Biology, 5e. New York: Garland Science.
3 Krebs, J., Goldstein, E.S., and Kilpatrick, S.T. (2018). Lewin's Genes XII. Burlington: Jones & Bartlett Learning.
2 Structure and Function of Cellular Macromolecules
Michael Wink
Heidelberg University, Institute of Pharmacy and Molecular Biotechnology (IPMB), Im Neuenheimer Feld 329, 69120 Heidelberg, Germany
In contrast to the diversity of life forms found in nature with several million species, the cells that make up all of these diverse organisms contain only a limited number of types of inorganic ions and molecules (Table 2.1). Among the most important macromoleculesof prokaryotic and eukaryotic cells are polysaccharides, lipids, proteins, and nucleic acids, which are constructed from comparatively few monomeric building blocks(Table 2.2). The membrane lipids(phospholipids, cholesterol) will also be considered in this context because they spontaneously form supramolecular biomembrane structures in the aqueous environment of a cell.
Table 2.1 Molecular composition of cells.
| Contents |
Bacterium (% of cell mass) |
Animal cell (% of cell mass) |
| Water |
70 |
70 |
| Inorganic ions |
1 |
1 |
| Small molecules (sugars, acids, amino acids) |
3 |
3 |
| Proteins |
15 |
18 |
| RNA |
6 |
1.1 |
| DNA |
1 |
0.25 |
| Phospholipids |
2 |
3 |
| Other lipids |
7 |
2 |
| Polysaccharides |
2 |
2 |
| Cell volume (ml) |
2 × 10 –12 |
4 × 10 –9 |
| Relative cell volume |
1 |
2000 |
Table 2.2 Formation and function of the cellular macromolecules.
| Basic building blocks |
Macromolecule |
Function |
| Simple sugar |
Polysaccharide |
Structural substances:composition of the cell walls (cellulose, chitin, peptidoglycan); constituents of connective tissues |
| Storage substances:starch, glycogen |
| Amino acids |
Protein |
Enzymes:important catalysts for anabolic and catabolic reaction processes |
| Hemoglobin:O 2and CO 2transport |
| Receptors:recognition of external and internal signals |
| Ion channels, ion pumps, transporters:transport of charged or polar molecules across biological membranes |
| Regulatory proteins:signal transduction through protein–protein interactions |
| Transcription regulators:regulation of gene activity |
| Antibodies:recognition of antigens |
| Structural proteins:structural organization of supramolecular complexes |
| Cytoskeleton:formation of molecular networks in the cell that are important for shape and function |
| Motor proteins:muscle contraction |
| Phospholipids, cholesterol |
|
Elements of biomembranes |
| Deoxynucleotide |
DNA |
Storage, replication, and safe transfer of genetic information; recombination |
| Nucleotide |
RNA |
rRNA:structural molecules for the construction of ribosomes |
| ribozymes and siRNA:catalytic and regulatory processes |
| tRNA:mediators in translation |
| mRNA:messengers and mediators between genes and proteins |
| snRNA:splicing of mRNA |
| snoRNA: chemically modify rRNA |
| siRNA: can influence gene expression by directing degradation of selective mRNAs and the establishment of compact chromatin structures |
| miRNA: can control gene activity,development, and differentiation by specifically blocking translation of particular mRNA |
| piRNA: bind to piwi proteins and protect germline from transposable elements |
| lncRNA: apparently play a role in regulating gene transcription |
Inorganic ions, sugars, amino acids, fatty acids, organic acids, nucleotides, and various metabolites are counted among the low‐molecular‐weight componentsand building blocks of the cell. The qualitative composition of cells is similar in prokaryotes and eukaryotes ( Table 1.1), even though eukaryote cells generally have a higher protein content and bacterial cells a higher RNA content. Animal cells have a volume that is 10 3times larger than that of bacterial cells.
Owing to their shared evolution, the structure and function of the important cellular molecules is very similar in all organisms, often even identical. Apparently, reliable and functional biomolecules were developed and, if useful for the producer, were selected early in evolution (Table 2.2) and are therefore still used today.
2.1 Structure and Function of Sugars
Monosaccharidesoccur in cells either as aldosesor as ketoses( Figure 2.1a). The most important monosaccharides have a chain length of three, five, and six carbon atoms and are called trioses, pentoses, and hexoses. Under physiological conditions, pentoses and hexoses can form ring structures through hemiacetal and hemiketal formation ( Figure 2.1b).
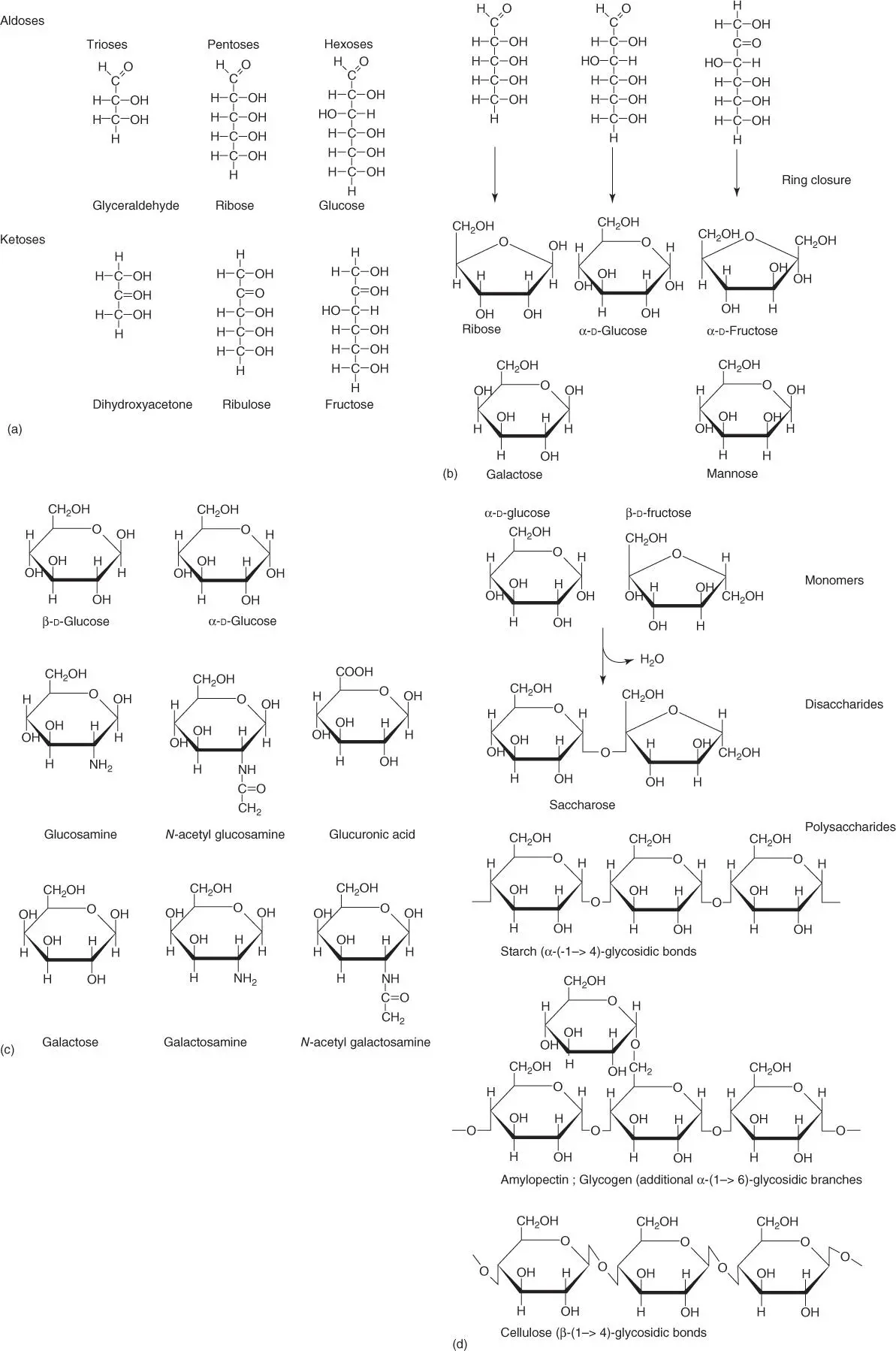
Figure 2.1 Composition and structure of sugar molecules. (a) Structures of the most important aldoses and ketoses. (b) Ring structures of pentoses and hexoses (hemiacetal and hemiketal formations), important isomers of glucose. (c) Important derivatives of glucose and galactose. (d) Formation of disaccharides and polysaccharides (starch [amylose], amylopectin, glycogen, cellulose).
Many important nitrogen‐containing derivatives of these monosaccharides ( Figure 2.1c) use galactose and glucose as a base. Examples include glucosamine, N‐acetylglucosamine, and glucuronic acid. These derivatives can be present either as glycosides or as part of a polysaccharide.
Condensation reactionsbetween sugar molecules result in the formation of glycosidic bondswith the elimination of a water molecule. As hydroxyl groups can be present in either the α or β position, the stereochemistry of sugar molecules is of great importance. The condensation of two sugar molecules results in the formation of a disaccharide( Figure 2.1d); that of three sugar molecules, correspondingly, is a trisaccharide. Oligosaccharidesare built from a few sugar monomers, and polysaccharides(e.g. starch, glycogen, cellulose, chitin, etc.) are made up of many sugar monomers.
Читать дальше




![Andrew Radford - Linguistics An Introduction [Second Edition]](/books/397851/andrew-radford-linguistics-an-introduction-second-thumb.webp)








