Table 1.2 Compartments of animal and plant cells and their main functions.
| Compartment |
Occurrence |
Functions |
| Nucleus |
A |
P |
Harbors chromosomes, site of replication, transcription, and assembly of ribosomal subunits |
| Endoplasmic reticulum (ER) |
|
|
|
| Rough ER |
A |
P |
Posttranslational modification of proteins |
| Smooth ER |
A |
P |
Synthesis of lipids and lipophilic substances |
| Golgi apparatus |
A |
P |
Posttranslational modification of proteins, modification of sugar chains |
| Lysosome |
A |
|
Harbors hydrolytic enzymes, degrades organelles and macromolecules, macrophages eat invading microbes |
| Vacuole |
|
P |
Sequestration of storage proteins, defense and signal molecules, contains hydrolytic enzymes, degrades organelles and macromolecules |
| Mitochondrium |
A |
P |
Organelle derived from endosymbiotic bacteria; contains circular DNA, own ribosomes; enzymes of citric acid cycle, β‐oxidation, and respiratory chain (ATP generation) |
| Chloroplast |
|
P |
Organelle derived from endosymbiotic bacteria; contains circular DNA, own ribosomes; chlorophyll and proteins of photosynthesis, enzymes of CO 2fixation and glucose formation (Calvin cycle) |
| Peroxisome |
A |
P |
Contains enzymes that generate and degrade H 2O 2 |
| Cytoplasm |
A |
P |
Harbors all compartments, organelles, and the cytoskeleton of a cell; many enzymatic pathways (e.g. glycolysis) occur in the cytoplasm |
A, animal; P, plant.
A highly resolved tree of life is based on completely sequenced genomes (Ciccarelli 2006). The image was generated using Interactive Tree Of Life (iTOL) (Letunic 2007), an online phylogenetic tree viewer and Tree of Life resource. Eukaryotes are colored red, archaea green, and bacteria blue.
The most important biochemical and cell biological charactersof Archaea, Bacteria, and Eukarya are summarized in Table 1.1.
As virusesand bacteriophages( Figure 1.3) do not have their own metabolism, they therefore do not count as organisms in the true sense of the word. They share several macromolecules and structures with cells. Viruses and bacteriophages are dependent on the host cells for reproduction, and therefore their physiology and structures are closely linked to that of the host cell.
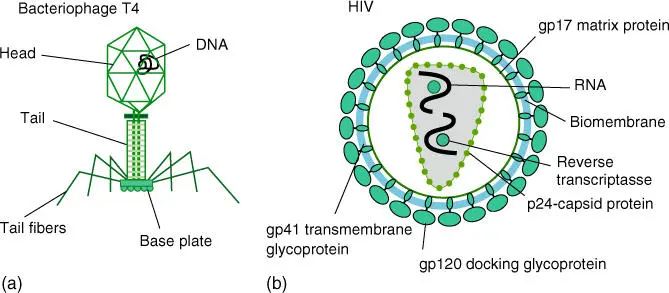
Figure 1.3 Schematic structure of bacteriophages and viruses. (a) Bacteriophage T4 and (b) structure of a retrovirus (human immunodeficiency virus causing AIDS).
Eukaryotic cells are characterized by compartmentsthat are enclosed by biomembranes (Table 1.2). As a result of these compartments, the multitude of metabolic reactions can run in a cell at the same time.
In the following discussion on the shared characteristics of all cells, the diverse differences that appear in multicellular organismsshould not be forgotten. The human body has more than 200 different cell types, which show diverse structures and compositions. These differences must be understood in detail if cell‐specific disorders, such as cancer, are to be understood and consequently treated. Modern technology with Next‐Generation Sequencing (NGS) allows a study of single cells at a genomic and transcriptomic level.
Before a detailed discussion of cellular structures and their functions (see Chapters 3– 5), a short summary of the biochemical basics of cellular and molecular biology is given in Chapter 2.
Progress in cell biology and biotechnology largely depends on innovative methods, as new methods often open windows to look deeper into biology and to solve old questions. Table 1.3summarizes some of the important tools, which are important for cell and molecular biology today.
Table 1.3 Important methodological tools of modern biology.
| Problem |
Technique/instrument |
Remarks |
| Structure elucidation of proteins |
Protein isolation, column chromatography (gel filtration, ion exchange, affinity) |
Chapter 7 |
|
Gel electrophoresis |
Chapter 7 |
|
Protein–protein interactions (FRET, two hybrid systems, FRAP) |
Chapters 19and 23 |
|
Crystallization |
|
|
X‐ray diffraction |
|
|
NMR |
|
|
Cryoelectron microscopy |
|
|
Mass spectrometry |
Chapter 8 |
|
Protein sequencing |
|
| DNA |
PCR and quantitative PCR (qPCR) |
Chapter 13 |
|
DNA/RNA isolation |
Chapter 9 |
|
DNA hybridization |
Chapter 11 |
|
Sanger sequencing |
Chapter 14 |
|
Restriction enzymes |
Chapter 12 |
|
Gel and capillary electrophoresis |
Chapter 10 |
|
Next generation sequencing |
Chapter 14 |
|
Microsatellite analysis |
Chapter 11 |
|
SNP analysis |
Chapters 14and 21 |
|
FISH |
Chapter 11 |
|
In situ hybridization |
Chapter 11 |
| RNA (transcriptomics) |
RNA‐seq (NGS) |
Chapters 14and 21 |
|
DNA microarrays |
Chapter 11 |
|
In situ hybridization |
Chapter 11 |
| Cell and tissue culture |
Cells with reporter genes |
|
|
Cell sorting |
|
|
Organoid cultures |
|
|
Stem cells |
|
|
Cancer cells |
|
|
Hybridoma cells for production of monoclonal antibodies |
|
|
Cell cycle analysis |
Chapter 18 |
|
Patch clamp recording |
Chapter 17 |
| Microscopy |
Light microscope (bright field, dark field, phase contrast, differential interference contrast) |
Chapter 19 |
|
Fluorescence microscope (confocal) |
Chapters 19and 20 |
|
Immunofluorescence and GFP fusion proteins |
Chapter 19 |
|
Super‐resolution microscopy (STED, SIM, PALM, STORM) |
Chapter 19 |
|
Atomic force microscopy |
Chapter 19 |
|
Electron microscope |
Chapter 19 |
|
Scanning electron microscope (SEM) |
|
|
Cryoelectron microscopy |
Chapter 19 |
|
Image processing |
|
| Cloning and expression |
Plasmid and viral vectors |
Chapter 15 |
|
Expression vectors |
Chapters 15and 16 |
|
Fermenters |
|
|
Genomic and cDNA libraries |
Chapter 21 |
|
Reverse genetics |
|
| Genetic engineering |
Transformation |
Chapter 15 |
|
Transfection |
Chapter 15 |
|
RNAi |
|
|
CRISPR–Cas gene editing |
|
|
Transgenic organism |
|
| New active agents |
Recombinant antibodies |
Chapter 16 |
|
Recombinant vaccines |
Chapter 16 |
|
Recombinant enzymes |
Chapter 16 |
| Information |
DNA sequences |
Chapter 24 |
|
Genomes |
Chapter 24 |
|
Proteins |
Chapter 23 |
|
System biology |
Chapter 23 |
Abbreviations: SNP, single nucleotide polymorphism; GFP, green fluoresecnt protein; NGS, next generation sequencing.
Читать дальше




![Andrew Radford - Linguistics An Introduction [Second Edition]](/books/397851/andrew-radford-linguistics-an-introduction-second-thumb.webp)








