The opposite process, endocytosis, also occurs continuously at the cytoplasmic membrane. In this process, vesicles bud off and migrate from the early endosomes to the late endosomesand finally deliver their contents to the lysosomes or the Golgi apparatus ( Figure 5.8).
The processes of endocytosisare subdivided:
Phagocytosis (uptake of microorganisms or dead cells).
Pinocytosis (uptake of liquids and smaller molecules).
Phagocytosis( Figure 5.8) is the function of the phagocytes (macrophages, neutrophils, and dendritic cells) of the cellular immune system. The phagocytosed cells are degraded in the lysosomes.
Pinocytosisis a continuous process: macrophages take up about 25% of their cell volume per hour via pinocytosis; in relation to the cytoplasmic membrane, this corresponds to a budding rate of the membrane in vesicles of 3% per minute. The surface of the membrane, which is taken up via endocytosis, corresponds to the cell surface, which is released via exocytosis( endocytosis–exocytosis cycle). During endocytosis the vesicles are filled with liquids and molecules that are present in the extracellular space. Through fluid‐phase endocytosispolar molecules can also enter the cell, which otherwise would not be taken up via diffusion or carriers.
An important variation of endocytosis is receptor‐mediated endocytosis. This is how lipoproteinssuch as low‐density lipoprotein ( LDL ) particles, which are loaded with cholesterol ester in blood, are recognized and bound by LDL receptorsof the target cell ( Figure 5.10). After binding, the clathrin‐coated endocytosis vesicle buds off and migrates via the endosomes to the lysosomes. There the receptors with exocytotic vesicles are returned to the cytoplasmic membrane, while the lipoproteins are degraded in the lysosome. The cholesterol esteris also cleaved by an esterase. Cholesterol is then available to the cell for synthesis or as a membrane lipid. Patients with defective LDL receptor genes(prevalence of 1 in 500) have an increased risk of myocardial infarction, as higher levels of cholesterol lead to arteriosclerosis.
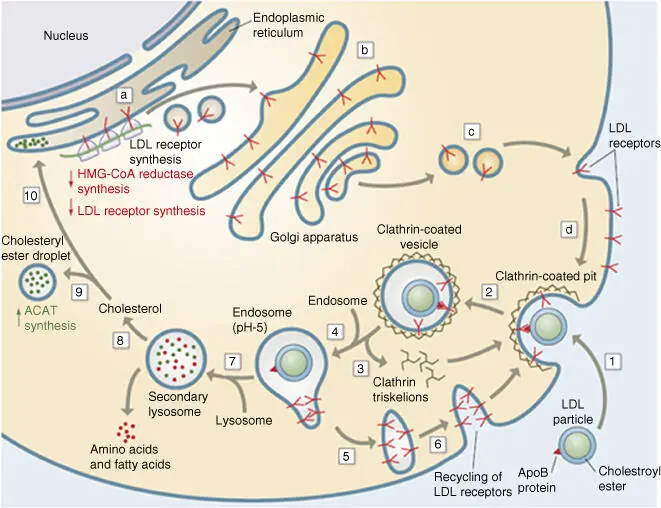
Figure 5.10 Schematic progression of receptor‐mediated endocytosis of LDL.
Source: From Voet et al. (2016).
1 Nigg, E.A. (1997). Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386: 779–787.
2 Voet, D., Voet, J.G., and Pratt, C.W. (2016). Fundamentals of Biochemistry, Live at the Molecular Level, 5e. Holboken, NJ: Wiley.
1 Alberts, B., Johnson, A., Lewis, L. et al. (2015). Molecular Biology of the Cell, 6e. New York: Garland Science.
2 Alberts, B., Bray, D., Hopkin, K. et al. (2019). Essential Cell Biology, 5e. New York: Garland Science.
3 Krebs, J., Goldstein, E.S., and Kilpatrick, S.T. (2018). Lewin's Genes XII. Burlington: Jones & Bartlett Learning.
6 Evolution and Diversity of Organisms
Michael Wink
Heidelberg University, Institute of Pharmacy and Molecular Biotechnology (IPMB), Im Neuenheimer Feld 329, 69120 Heidelberg, Germany
Molecular cell biologyfocuses more and more on model organisms. Examples are, among others, the Gram‐negative bacteria Escherichia coli , the brewer's yeast ( Saccharomyces cerevisiae ), the nematode worm ( Caenorhabditis elegans ), the fruit fly ( Drosophila melanogaster ), the common mouse ( Mus musculus ), the zebra fish ( Danio rerio ), the human ( Homo sapiens ), and wall cress ( Arabidopsis thaliana ) as a representative of higher plants. Extensive knowledge about molecular and cellular biology was gained from these model organisms. As these organisms share a common evolutionary history, it is assumed that basic characteristics found in one model organism are also valid for all other organisms. This can, but must not always, be true. In some cases nature has also found different solutions to the same problem ( convergent evolution). Nevertheless, we see an astonishing degree of similarity in fundamental mechanisms and structures.
However, there is also an astonishing diversity at the organismal level. In the future of biotechnology, we must not ignore the diversity of organisms, with presently known 2 million species (perhaps over 10 million species exist) and complex adaptations. Many of them offer evolutionarily derived solutions for problems that are of great interest for biotechnological purposes or applications.
Figure 6.1shows a tree of lifereconstructed over nucleotide sequences of 31 genes from 191 species whose genomes were completely sequenced (Ciccarelli et al. 2006). This tree illustrates the lines of development for the different kingdoms. Within prokaryotes, two large domains are recognizable: the Bacteria(or simply bacteria) and the Archaea(or archaebacteria). Important biochemical differences were already summarized in Table 1.1.
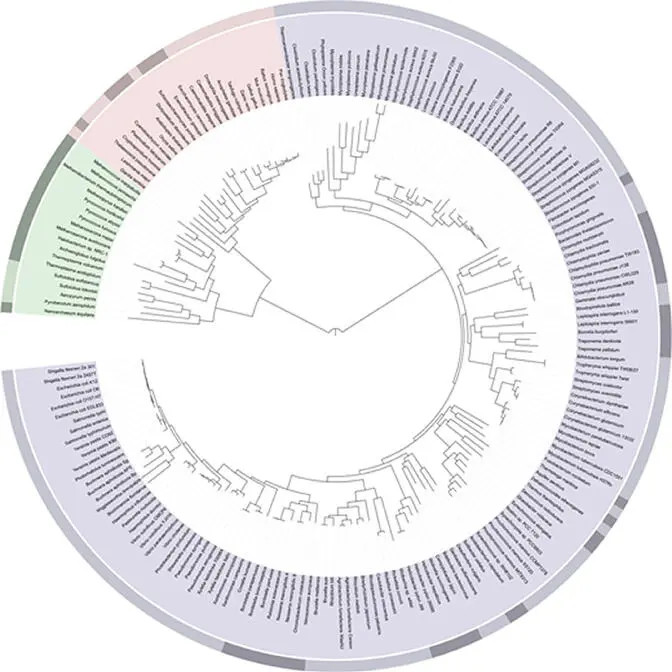
Figure 6.1 A phylogenetic tree of life, showing the relationship between species whose genomes had been sequenced as of 2006. The very center represents the last universal ancestor of all life on earth. The different colors represent the three domains of life: pink represents eukaryota (animals, plants, and fungi), blue represents bacteria, and green represents archaea. Note the presence of Homo sapiens (humans) second from the rightmost edge of the pink segment. The light and dark bands along the edge correspond to clades: the rightmost light red band is Metazoa, with dark red Ascomycota to its left, and light blue Firmicutes to its right.
Source: https://commons.wikimedia.org/wiki/File:Tree_of_life_SVG.svg..
The evolution of the ancestor of a eukaryotic celland the uptake of bacteria (endosymbiotic origin of mitochondria and chloroplasts) were a key innovation of early evolution. While the incorporation of mitochondria only occurred once in evolution, there is reasonable evidence for the assumption that the incorporation of cyanobacteria (leading to chloroplasts) occurred many times (especially within different groups of algae).
There are large differences in cellular structure and function between prokaryotes and eukaryotes. Table 1.1summarized the important characteristics. The eukaryotic cell is distinctly further developed (Figure 1.2) and is able to carry out different processes at the same time in a single cell. This required the development of separated reaction spaces – cellular compartments(Table 1.2) – in the early stages of evolution.
A simplified overview of the origin of organisms is shown in Figures 1.1and 6.1. Due to lack of space, it is not possible to go into more detail for the different organisms in the specific individual domains of the living kingdoms. To give biotechnologists a quick orientation about which organism they are focusing on and where these organisms stand in the tree of life, a short systematic synopsis of the organisms is put together in the following. For simplicity, only the large groups of protists (Table 6.1; Figure 6.2), plants (Table 6.2; Figure 6.3), and animals (Table 6.3; Figures 6.4and 6.5) will be more closely characterized (a good short overview can be found in Campbell et al. (2018)). Apparently, the protozoa do not form a monophyletic clade as formerly assumed, but several independent evolutionary lineages. Traditionally, algae, and sometimes even fungi and bacteria, have been included in plants. As can be seen from Figures 6.1and 6.2, only the metabionta with red algae, green algae, and land plants forms a monophyletic unit. Fungi cluster with Opisthokonta and thus much closer to animals and then to plants. Among animals, the Protostomia have now been separated in Ecdysozoa and Lophotrochozoa on account of molecular and anatomical data ( Figure 6.4). According to the rules of cladistics, only monophyletic groups should be accepted. This requires a restructuring of some of the groups of organisms that had been grouped together, such as protists, mosses, fishes, and reptiles (Lecointre and Le Guyader 2007).
Читать дальше





![Andrew Radford - Linguistics An Introduction [Second Edition]](/books/397851/andrew-radford-linguistics-an-introduction-second-thumb.webp)








