5.1 Import and Export of Proteins via the Nuclear Pore
The cell nucleus is enclosed by a nuclear envelope consisting of two concentric membranes (see Section 3.1.2). Every nuclear envelope contains over 3000–4000 NPCs. The NPC in animals has a molecular weight of 125 million Da and is made up of 30 proteins, which are termed nucleoporins. An NPC consists of 500–1000 individual proteins. NPCs are able to import (e.g. histone proteins, DNA polymerases, RNA polymerases, transcriptional regulators, telomerase, RNA processing enzymes) or export (e.g. the subunits of the ribosomes that are assembled in the nucleolus, all RNAs) a large number of proteins in a short time. About 1000 macromolecules per second pass a single NPC. The nuclear pores are filled with water and allow substances smaller than 5000 Da to pass through unhindered. For larger molecules (>60 000 Da), they are highly selective. Cargo proteinsmust bear the correct signal sequence (Table 5.1). The structure of a nuclear pore is schematically represented in Figure 5.2.

Figure 5.2 Structure of a nuclear pore (reconstructed from electron microscopy images). The nuclear pore complex contains about 30 different proteins. Inner diameter = 9 nm. The upper side is oriented toward the cytosol.
Source: Nigg (1997). Reproduced with permission of Springer Nature.
For the import or export, mobile nuclear import receptors (importins )and export receptors( exportins) are required. These receptors must, on the one hand, recognize the recognition signal of the protein to be transported ( cargo protein; Table 5.1) and, on the other hand, interact with the nucleoporins of the nuclear pores. Nuclear import and export are shown schematically in Figure 5.3. First, a cargo proteinand nuclear import receptor complexare formed. As soon as a cargo protein/import receptor complex has arrived at the inner side of the nuclear membrane, a guanosine triphosphate ( GTP )‐ binding protein( Ran‐GTP) binds to the import receptor. A conformational change occurs, and a cargo protein is released. The complex of Ran‐GTP and the import receptor binds to nucleoporin and transports it through the pore in the direction of cytosol. Once it arrives, Ran‐GTP is dephosphorylated and dissociated from the import receptor as Ran‐ guanosine diphosphate ( GDP ), whereby the receptor is reactivated. Export out of the nucleus occurs with a similar principle ( Figure 5.3). The change from Ran‐GTP to Ran‐GDP is catalyzed by a GTPase‐activating protein( GAP); the exchange of GDP to GTP in the nucleus is assisted by a guanine exchange factor ( GEF ). The export of cargo from the nucleus to the cytosol follows a similar logic ( Figure 5.3).
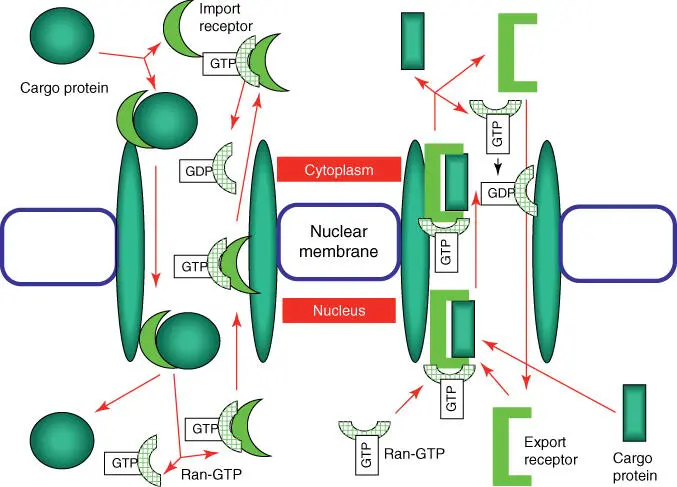
Figure 5.3 Simplified model of the import and export of proteins via the nuclear pore. The left side represents protein import via a nuclear pore; the right side represents the export of cargo proteins.
5.2 Import of Proteins in Mitochondria, Chloroplasts, and Peroxisomes
Proteins that should function inside the mitochondriaor chloroplastsare synthesized as precursor proteinson cytosolic ribosomes and carry a recognition sequence on the N‐terminal(Table 5.1). After uptake by the organelle, this signal sequence is removed by a signal peptidase. The import progresses via a multienzyme complex: the translocase of outer membrane ( TOM ) complexbinds a precursor protein and transports it over the outer mitochondrial membrane. Further transport over the inner mitochondrial membrane is taken over by TIM22and TIM23complexes ( Figure 5.4). When membrane proteins are imported, they contain an additional signal sequence, which is then recognized by the OXA complex. The OXA complex ensures that membrane proteins, whether synthesized by the mitochondria or imported out of the cytosol, are incorporated correctly in the inner mitochondrial membrane. A SAM complexhelps to place proteins in the outer mitochondrial membrane.
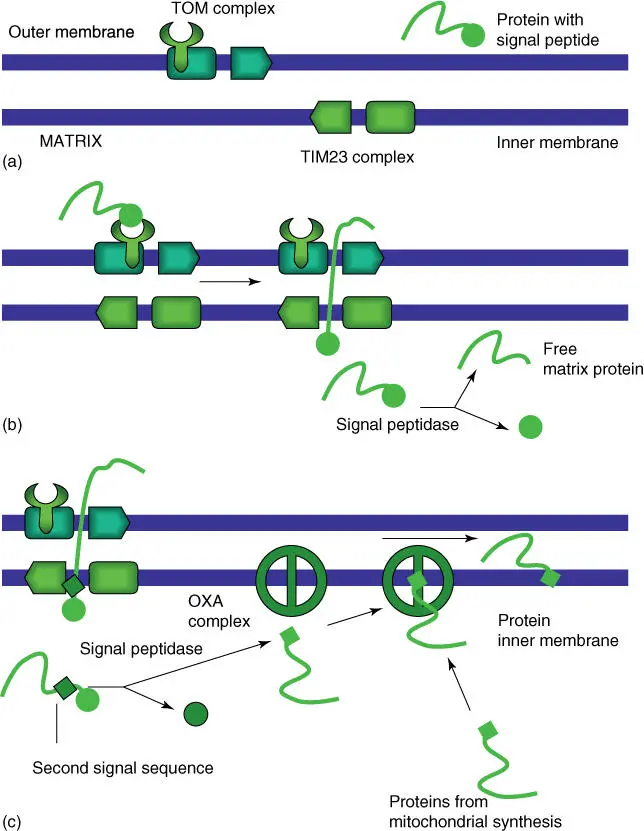
Figure 5.4 Schematic overview of the uptake of a precursor protein by the mitochondria and the assembly of membrane proteins in the inner mitochondrial membrane. Eukarya, translocase of outer membranes; TIMs, translocase of inner membranes. (a) Setup of transport systems. (b) Cooperation between Eukarya and TIM complexes. (c) Function of the OXA complex.
Transporting precursor proteins into chloroplastsfollows a similar scheme. A second signal is necessary for transport into the thylakoid.
Peroxisomesharbor enzymes to break down hydrogen peroxide (catalase) and the enzymes of β‐oxidation of fatty acids. Proteins targeted for peroxisomes carry a short signal peptide of three amino acids (Ser‐Lys‐Leu) on their C‐terminus. Peroxisomes carry a complex of protein translocators, peroxins(e.g. Pex1, Pex5, Pex6, Pex7), which are activated by adenosine triphosphate (ATP).
5.3 Protein Transport into the Endoplasmic Reticulum
In electron microscope photographs, the rough ERis recognized by its large number of ribosomes, which look as if they are tightly bound to the ER membrane (Figure 1.2). These ribosomesare in the process of synthesizing proteins, which are then secreted into the ER lumen. These proteins are characterized by a specific signal peptide in the N‐terminal(Table 5.1).
In principle, protein biosynthesis begins on the free ribosomes in the cytoplasm. When a protein exhibiting an ER import signal peptideis synthesized, a signal recognition particle ( SRP )will bind to the signal sequence. In the next step, SRP binds an SRP receptorpresent at the ER membrane and therefore brings the translating ribosome into the vicinity of a protein translocator (consisting of Sec61, 62, 63, 71, and 72). Figure 5.5schematically shows the import of a protein into the ER lumen. As soon as a protein is completely synthesized and the C‐terminal of the protein has arrived in the ER lumen, a signal peptidasecleaves the signal recognition sequence, and the protein is freed into the ER lumen.
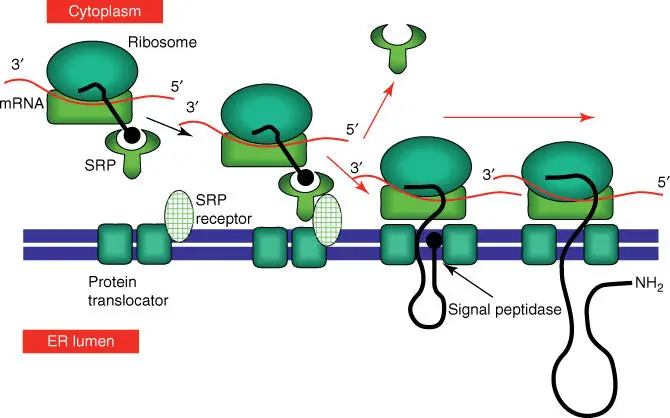
Figure 5.5 Simplified scheme of the import of a protein into the ER lumen.
Читать дальше







![Andrew Radford - Linguistics An Introduction [Second Edition]](/books/397851/andrew-radford-linguistics-an-introduction-second-thumb.webp)








