The widely universal triplet code has a specific start signal. Since methionine(in eukaryotes) and N‐formylmethionine(in bacteria and chloroplasts) are the first amino acids to be built into polypeptides, the universal start codonis AUG(far more seldom, GUG is present). In most cases, however, methionine is removed by specific proteases following translation. When the start of the translation shifts only one or two nucleotides, resulting in a shift of the reading frame( frameshift) ( Figure 4.13), a totally new protein results. This means that the start codon must be strictly preserved in order to produce reproducible proteins. In animal(but not in plant) mitochondria, there is a deviation from the universal genetic code (e.g. AUA is used for translation initiation and codes for methionine). However, in eukaryotic ribosomes this codon codes for isoleucine; AGG/A is used as a termination codon by vertebrate mitochondria, while it usually codes for arginine. UGA, which is usually a stop codon, codes for tryptophan in animal mtDNA.
Usually the codons that code for the same amino acid differ in the third codon position. Every codon is recognized by tRNA via the anticodon sequence. Within the so‐called degenerate codonsthat all code for the same amino acid, usually only one tRNA exists, one which tolerates a mismatchingin the third codon position. Overall, about 31 tRNAs have been discovered in the eukaryotic system and 22 tRNAs in mitochondria.
4.3 Protein Biosynthesis (Translation)
Protein biosynthesistakes place in ribosomes– intricately constructed multienzyme complexes in which different rRNAs play an important role ( Figure 4.22). rRNAsbelong to the most prevalent macromolecules of the cell. The numerous copies of the rDNA cassettes in the genome ( Figure 4.21) indicate that this gene must be transcribed very often in order to produce the large number of rRNA molecules that every cell requires. Just for E. coli alone, the number of rRNA molecules is estimated to be 38 000. In a mammalian cell more than 1 million rRNA copies exist. The rRNA genes for 18S, 5.8S, and 26S rRNAare transcribed by RNA polymerase I together, and the individual rRNAs are produced afterward by splicing. Nucleotides of the precursor RNA are chemically modified by snoRNAsbefore splicing (Figure 2.20).
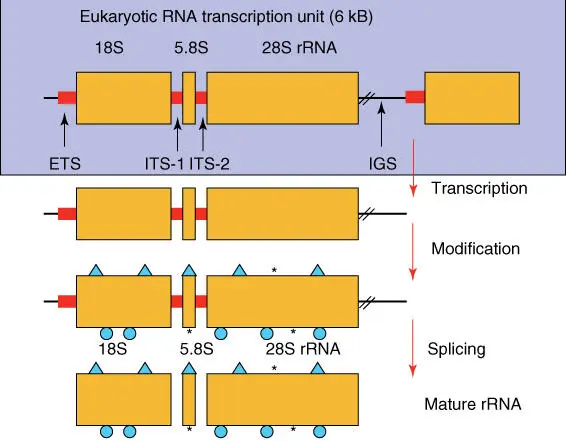
Figure 4.21 Structure of RNA cassettes and synthesis of rRNA. ITSs, internal transcribed spacers; IGSs, intergenic spacers; 5S rRNA genes are transcribed separately.
Figure 4.22shows the assembled building blocks of prokaryotic and eukaryotic ribosome s. As mitochondriaand chloroplastscontain their own ribosomes, which originated from bacteria (see Section 3.1.3), the expected type of rRNAs corresponds to those of bacteria (note that in mitochondria a 12S rRNA is present instead of the 23S rRNA).
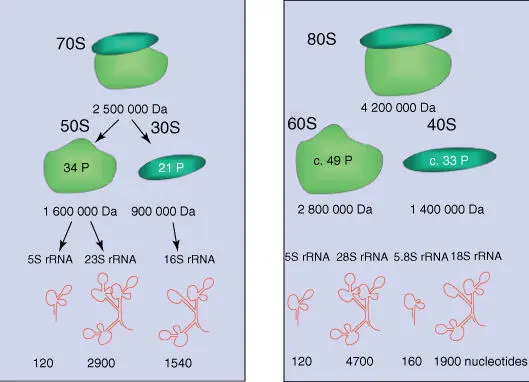
Figure 4.22 Structure of (a) prokaryotic and (b) eukaryotic ribosomes. For the structure of rRNA, see Figure 2.20.
16/18S rRNA and 23/28S rRNAs exhibit complex spatial structures, which are conserved over a wide range of organisms (Figure 2.20). Even though the RNAs consist of single strands, they form complementary double strands(so‐called stem structures) at many sites in aqueous environments. The nucleotide sequence of stem structures is very strongly preserved in evolution. The situation is different for the loops, in which the nucleotides have been modified posttranscriptionally. This phenomena of base modificationis especially observed with tRNAs (but also in rRNAs), in which more than 50 modified nucleotides have been discovered. Substituted basesare thiouracil, 5‐methylcytosine, dihydrouracil, thiothymine, thiocytosine, N 4‐acetylcytosine, 1‐methylhypoxanthine, 1‐methylguanine, and N 6‐methyladenine. There are comparatively many substitutions, deletions, insertions, and inversions present in the loops. Before NGS, genetic trees of all organisms have been reconstructed from the nucleotide sequences of the conserved rRNAs, giving them a special role in molecular evolution. The tree of lifeand the classification of species were largely based on the analysis of conserved rDNA genes. Today, because of the wide availability of NGS, such trees are often reconstructed from partial genomes (see Chapter 1).
The ribosomal proteins are arranged around the rRNA, together constituting a complex nanomachine known as the ribosome(Figures 4.22and 4.23). Both ribosomal subunits are assembled in the cell nucleolusand are transported individually into the cytosol through the nuclear pores. Free mRNA molecules are recognized by the small subunits, which are first loaded with methionine tRNAand guanosine triphosphate (GTP)‐activated initiation factors( eIF‐2). The small subunit slides along the mRNA until the first start codon AUGis reached, where methionine tRNA is bound via its anticodon UTC. Following the dissociation of the initiation factor eIF‐2, the large ribosomal subunit is able to bind, and the ribosome is positioned ready to begin translation. There are three formally distinguished binding sites: the arriving aminoacyl‐tRNAsbind to the A‐site, the tRNA with the peptide chain sits in the P‐site, and the E‐sitereleases the free tRNA after peptide transfer ( Figure 4.23).
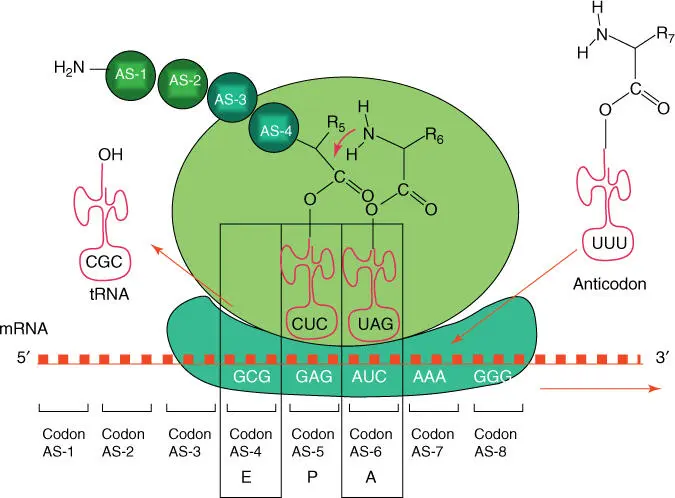
Figure 4.23 Schematic illustration of protein biosynthesis in ribosomes. Three binding sites are distinguished in ribosomes: E, P, and A.
In the A‐site, the arriving aminoacyl‐tRNAs (loaded with amino acids) are hybridized via their anticodon to the corresponding triplet codon on the mRNA ( Figure 4.24). In the next step, the peptide residue on the tRNA in the P‐site is transferred to the aminoacyl‐tRNA in the A‐site (peptidyl transfer is catalyzed by the rRNA; Figure 4.25). Next, the ribosome moves along three nucleotides on the mRNA and releases the free tRNA from the P‐site, which now carries the tRNA with the growing peptidyl residue. These steps are repeated until a stop codonis reached. A specific release factorthen binds and blocks access for further aminoacyl‐tRNAs to the A‐site. As a consequence, the peptide chain is released.
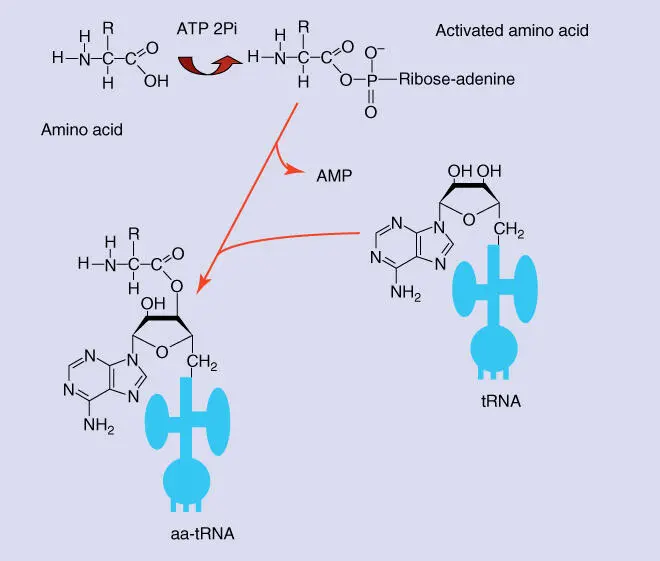
Figure 4.24 Loading tRNA with an amino acid. First the amino acid is activated through the binding of ATP. The activated amino acid is transferred to the 3′‐OH group of the terminal adenine residue of the tRNA, and an adenosine monophosphate (AMP) residue is set free. This reaction is catalyzed by aminoacyl‐tRNA synthetase that is specific for every amino acid. aa‐tRNA, aminoacyl‐tRNA (i.e. a tRNA loaded with an amino acid).
Читать дальше







![Andrew Radford - Linguistics An Introduction [Second Edition]](/books/397851/andrew-radford-linguistics-an-introduction-second-thumb.webp)








