Owing to the redundant genetic code, not every point mutation in a gene leads to a change in the amino acid sequence. Twenty‐five percent of all theoretically possible substitutions are synonymous, 4% lead to stop codons, and 71% to amino acid exchange. Nucleotide substitutions in the third codon position do not lead to a change in the amino acid in about 69% of cases (referred to as a silent mutation). Should deletions or insertions occur within a coded sequence, a frameshift mutation results, which almost always leads to severe damage of the corresponding proteins ( Figure 4.13).
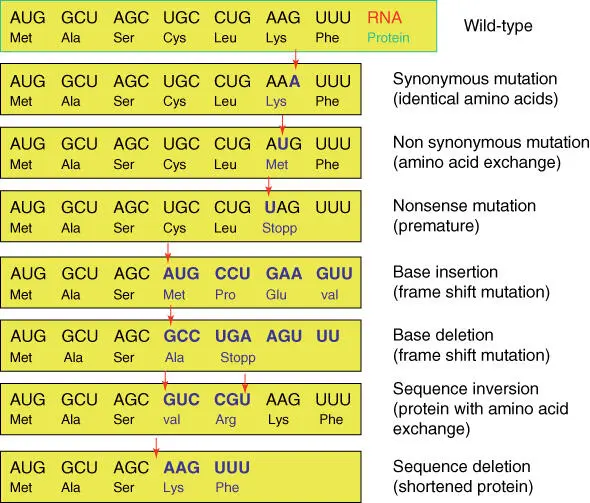
Figure 4.13 Consequences of gene mutations.
Point mutationsthat cause amino acid exchange (nonsynonymous substitution) often have a negative effect on the corresponding proteins. If the mutation is in the active site or in a binding site, a total loss of function can result. As diploid organisms have at least two copies of every gene, positioned on autologous chromosomes, such a point mutation usually does not lead to physical damage if the other copy of the gene is still intact. Only after both copies have been damaged is there a loss of the corresponding protein function ( Figure 4.14). Such disruption is often the basis for disease. This is especially true when the mutation arises in gametes and is then inherited. If the disorder is only in one allele, then it is referred to as a heterozygote character. If both alleles are identical, this is known as a homozygote character( Figure 4.14). In certain genes, such point mutations and the consequences on the health of the individual are widely known. They are referred to as SNPs (single nucleotide polymorphisms ),if they occur in several individuals of a population. One of the most important tasks for molecular biotechnologists is the development of diagnostic systems to quickly and reproducibly detect such SNPs. For this purpose mass spectrometry, DNA sequencing, NGS, PCR methods, and DNA chip strategies can be used (see Chapters 13and 14). This information can help to rationally treat diseases and can lead to a better understanding of their causes.
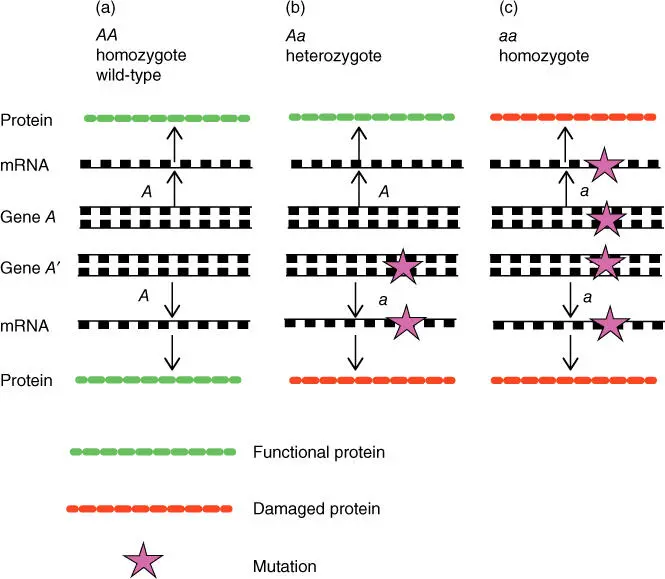
Figure 4.14 Inheritance of mutations leading to the loss of protein function. Gene A codes for a functional protein; gene a codes for a protein rendered functionless by a mutation. (a) Wild‐type genotype AA . (b) heterozygote genotype Aa . (c) homozygote recessive genotype aa .
4.2 Transcription: From Gene to Protein
Originally, mutation and recombination units were regarded as genes; in the 1950s the “one gene, one protein”hypothesis was developed ( DNA makes RNA, which makes proteins). Today, the gene is defined as a transcription unit. In the meantime, the intron/exon structure and the noncoding regulatory sequences, which also belong to the gene, have been recognized. Since mRNAs can be alternatively spliced, the statement “one gene, one protein” is no longer true in the strictest sense. The genetic information flows in all organisms from the gene to the mRNA and to the protein ( Figure 4.15). Only retroviruses can reversely translate RNA into DNA using a reverse transcriptase, but in no case has a translation of the amino acid sequence of a protein into a nucleotide sequence been shown.
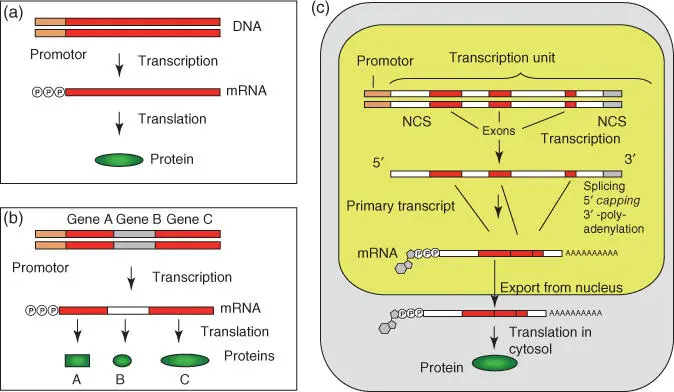
Figure 4.15 From gene to protein: comparison of prokaryotes and eukaryotes. (a) Simple prokaryotic gene: the mRNA is translated to a protein. (b) Bacterial operon: the primary transcript holds the genetic information for many genes (polycistronic mRNA). In protein biosynthesis, the protein units are synthesized separately. (c) Eukaryotic system: in the nucleus a primary transcript from RNA polymerase II is synthesized from which the intron regions are removed in preceding steps. At the 5′‐end a 7‐methylguanosine cap is added and a poly(A) tail is added to the 3′‐end. The completed mRNA is transported through the nuclear pore complex into the cytosol where it is translated into proteins by the ribosomes. NCS, noncoding sequence.
In eukaryotes, three different RNA polymerasesexist, which transcribe DNA into mRNA (plus small nucleolar RNA [snoRNA], miRNA, siRNA, lncRNA, most small nuclear RNA [snRNA]) ( RNA polymerase II), ribosomal RNA (rRNA) ( RNA polymerase I), or into other functional RNAs (e.g. tRNAs, 5S rRNA, snRNA, other small RNAs; RNA polymerase III). In prokaryotes, only one RNA polymerase is present. The translation of DNA into RNA is termed transcription.
As with replication, in transcription the DNA double helix is locally unwound, so that the RNA polymerase can synthesize the RNA (mRNA, rRNA, tRNA, and other RNAs) complementary to the template DNA strand( Figure 4.16). The DNA strand bearing an identical sequence to the mRNA (except that the T has been replaced with U) is referred to (in a confusing manner) as the coding strand. In addition, the sequence of the coding strand is written in the 5′ → 3′ orientation and is also stored in this format in sequence data banks.
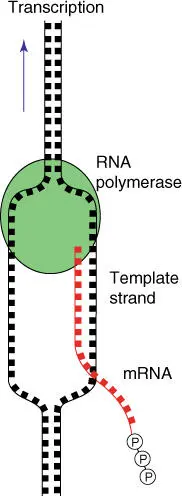
Figure 4.16 Schematic overview of the function of RNA polymerase and transcription.
| Coding strand |
5′‐ GGC TCC CTA TTA GCA GTC TGC CTC ATG ACC‐3′ |
| Template strand |
3′‐ CCG AGG GAT AAT CGT CAG ACG GAG TAC TGG‐5′ |
| mRNA |
5′‐ GGC UCC CUA UUA GCA GUC UGC CUC AUG ACC‐3′ |
The bacterial RNA polymeraseis a multienzyme complex containing a removable sigma factor. The sigma factor recognizes promoter regions of genes and assists the RNA polymerase in finding the transcription start. In Escherichia coli , the promoter is made up of two hexamer sequence motives, which are positioned 10 or 35 bases in front of a gene. The consensus sequences are …TTGACA…TATAAT…. Prokaryotic genes are usually organized in the form of operons( Figure 4.15b): genes that belong together, such as those that code for enzymes of a biosynthesis pathway, lie beside one another, and are controlled by a common promoter, which consists of an operatoras the control element ( Figure 4.17a). Well‐known examples include the Lac operonand tryptophan operon in bacteria, which are regulated by transcriptional activators and transcriptional repressors. These operons are important tools in biotechnology to control the expression of recombinant genes.
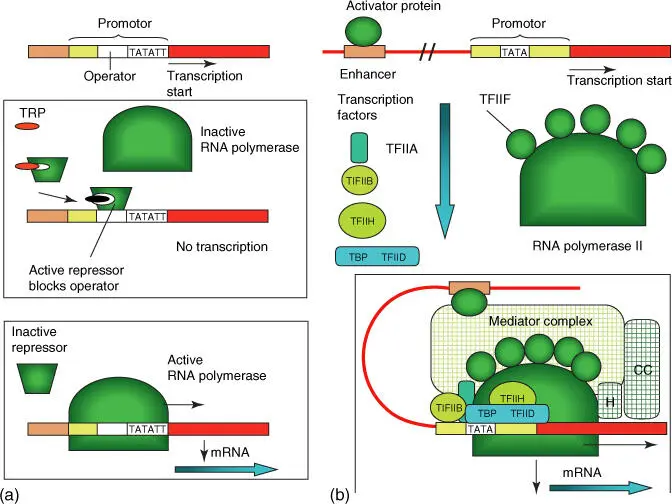
Figure 4.17 Simplified schematic illustration of the control of gene expression in prokaryotes and eukaryotes. (a) Bacteria: example tryptophan operon. When the amino acid tryptophan (TRP) is available in excess, the transcription of tryptophan biosynthesis enzymes is then inhibited by a repressor that is activated through the tryptophan, blocking the operator in the promoter. If no tryptophan is available, then the repressor dissociates from operator, and RNA polymerase can begin with transcription (bottom illustration). (b) Eukaryotes: transcription can only begin when an activated protein has bound to the enhancer and the complete transcription factors (Table 4.6) form a transcription complex together with the RNA polymerase II. The connections between the activator protein and the transcription complex are established through a mediator protein, which collaborates with a chromatin remodeling complex (CC) and a histone‐modifying enzyme (H). In addition, proteins are present that dissolve nucleosome complexes so that the DNA is accessible to the RNA polymerase.
Читать дальше








![Andrew Radford - Linguistics An Introduction [Second Edition]](/books/397851/andrew-radford-linguistics-an-introduction-second-thumb.webp)








