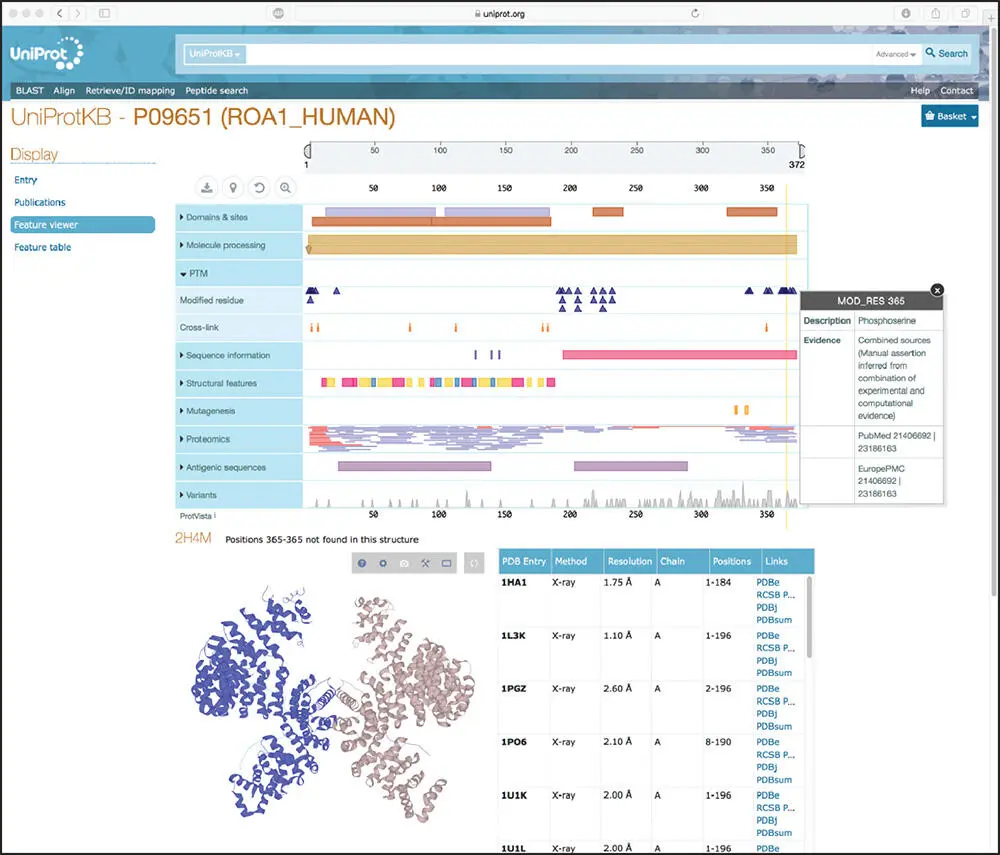
Figure 1.4 The Feature viewer rendering of the record for the human heterogeneous nuclear ribosomal protein A1 within UniProtKB. Clicking the Display link, found in the upper left portion of the window, provides access to the Feature viewer. Any of the sections can be expanded by clicking on the labels in the blue boxes to the left of the graphic. See text for details.
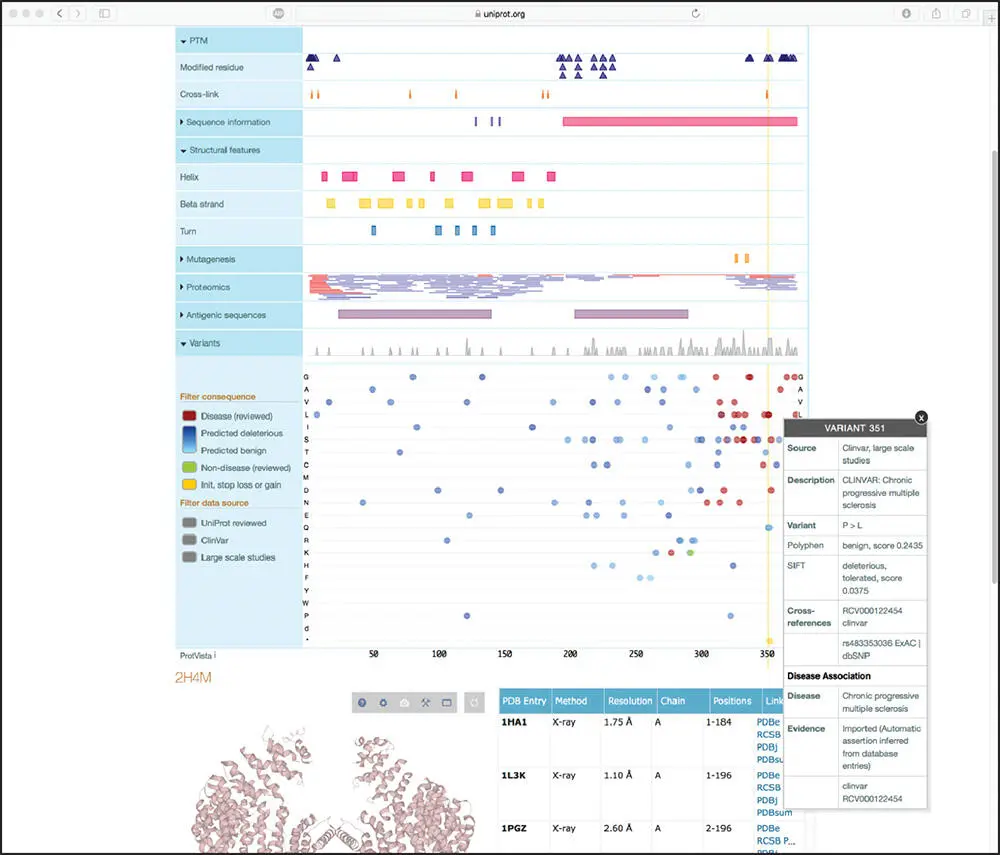
Figure 1.5 Expanding the PTM, Structural features, and Variants sections within the Feature viewer display shows the position of all post-translational modifications (PTMs), alpha helices, beta strands, and beta turns within the human heterogeneous nuclear ribosomal protein A1, as well as the location of putatively clinically relevant point mutations. Clicking on any of the variants produces a pop-up window with additional information; here, the pop-up window provides disease association data for the proline-to-leucine variant at position 351 of the sequence. See text for details.
The rapid pace of discovery in the genomic and proteomic arenas requires that databases are built in a way that facilitates not just the storage of these data, but the efficient handling and retrieval of information from these databases. Many lessons have been learned over the decades regarding how to approach critical questions regarding design and content, often the hard way. Thus, the continued development of currently existing databases, as well as the conceptualization and creation of new types of databases, will be a critical focal point for the advancement of biological discovery. As should be obvious from this chapter, keeping databases up to date and accurate is a task that requires the active involvement of the biological community ( Box 1.3). Therefore, it is incumbent upon all users to ensure the accuracy of these data in an active fashion, engaging the curators in a continuous dialog so that these widely used resources continue to remain a valuable resource to biologists worldwide.
Box 1.3Ensuring the Continued Quality of Data in Public Sequence Databases
Given the roles of DDBJ, EMBL, and GenBank in maintaining the archive of all publicly available DNA, RNA, and protein sequences, the continued usefulness of this resource is highly dependent on the quality of data found within it. Despite the high degree of both manual and automated checking that takes place before a record becomes public, errors will still find their way into the databases. These errors may be trivial and have no biological consequence (e.g. an incorrect postal code), may be misleading (e.g. an organism having the correct genus but wrong species name), or downright incorrect (e.g. a full-length mRNA not having a CDS annotated on it). Sometimes, records may have incorrect reference blocks, preventing researchers from linking to the correct publication describing the sequence. Over time, many have taken an active role in reporting these errors but, more often than not, these errors are left uncorrected.
While the individual INSDC members have the responsibility for hosting and disseminating the data found within their databases, keep in mind that the ownership of the data rests with the original submitter – and these original submitters (or their designees) are the only ones who can make updates to their database records. To keep these community resources as accurate and up to date as possible, users are actively encouraged to report any errors found when using the databases in the course of their work so that the database administrators can follow up with the original submitters as appropriate.
Given below are the current e-mail addresses for submitting information regarding errors to the three major sequence databases. As all the databases share information with each other nightly, it is only necessary to report the error to one of the three members of the consortium. Authors are actively encouraged to check their own records periodically to ensure that the information they previously submitted is still accurate. Even though this charge to the community is discussed here in the context of the three major sequence databases, all databases provide similar mechanisms through which incorrect information can be brought to the attention of the database administrators.
| DDBJ |
ddbjupdt@ddbj.nig.ac.jp |
| EMBL |
datasubs@ebi.ac.uk |
| GenBank |
gb-admin@ncbi.nlm.nih.gov |
As alluded to above, the range of publicly available data obviously goes well beyond human data, whether sequence based or not. As the major public sequence databases need to be able to store data in a fairly generalized fashion, these databases often do not contain more specialized types of information that would be of interest to specific segments of the biological community. To address this, many smaller, specialized databases have emerged and have been developed and curated by biologists “in the trenches” to fulfill specific needs. These databases, which contain information ranging from strain crosses to gene expression data, provide a valuable adjunct to the more visible public sequence databases, and users are encouraged to make intelligent use of both types of databases. An annotated list of such databases can be found in the yearly Database issue of Nucleic Acids Research (Rigden and Fernández 2018).
The position of this chapter at the beginning of this book reflects the belief that an understanding of biological databases is the first step toward being able to perform robust and accurate bioinformatic analyses. The reader is very strongly encouraged to take the time to understand the structure of the data found within these databases, as the basis for finding sequence data of interest and performing the more advanced analyses described in the chapters that follow.
The author thanks Rolf Apweiler for the use of material from the third edition of this book.
| DDBJ Database Divisions |
www.ddbj.nig.ac.jp/ddbj/data-categories-e.html |
| DNA Database of Japan (DDBJ) |
www.ddbj.nig.ac.jp |
| EMBL Nucleotide Sequence Database |
www.embl.org |
| ENA Data Formats |
www.ebi.ac.uk/ena/submit/data-formats |
| European Bioinformatics Institute |
www.ebi.ac.uk |
| GenBank |
www.ncbi.nlm.nih.gov |
| GenBank Database Divisions |
www.ncbi.nlm.nih.gov/genbank/htgs/divisions |
| Genome Ontology Consortium |
geneontology.org |
| INSDC Feature Table Definition |
insdc.org/documents/feature_table.html |
| International Society for Biocuration |
biocuration.org |
| NCBI Data Model |
www.ncbi.nlm.nih.gov/IEB/ToolBox/SDKDOCS/DATAMODL.HTML |
| NCBI Protein Database |
www.ncbi.nlm.nih.gov/protein |
| Nucleic Acids Research Database issue |
academic.oup.com/nar |
| Protein Data Bank (PDB) |
www.rcsb.org |
| Protein Identification Resource (PIR) |
pir.georgetown.edu |
| Protein Research Foundation |
www.proteinresearch.net |
| RefSeq |
www.ncbi.nlm.nih.gov/refseq |
| Swiss-Prot (EBI) |
www.ebi.ac.uk/uniprot |
| Swiss-Prot (ExPASy) |
web.expasy.org/docs/swiss-prot_guideline.html |
| UniProt Consortium |
www.uniprot.org |
Читать дальше






