FT CDS join(201..224,1550..1920,1986..2085,2317..2404,2466..2629) FT /codon_start=1 FT /gene="eIF4E" FT /product="eukaryotic initiation factor 4E-II" FT /note="Method: conceptual translation with partial peptide FT sequencing" FT /db_xref="GOA:P48598" FT /db_xref="InterPro:IPR001040" FT /db_xref="InterPro:IPR019770" FT /db_xref="InterPro:IPR023398" FT /db_xref="PDB:4AXG" FT /db_xref="PDB:4UE8" FT /db_xref="PDB:4UE9" FT /db_xref="PDB:4UEA" FT /db_xref="PDB:4UEB" FT /db_xref="PDB:4UEC" FT /db_xref="PDB:5ABU" FT /db_xref="PDB:5ABV" FT /db_xref="PDB:5T47" FT /db_xref="PDB:5T48" FT /db_xref="UniProtKB/Swiss-Prot:P48598" FT /protein_id="AAC03524.1" FT /translation="MVVLETEKTSAPSTEQGRPEPPTSAAAPAEAKDVKPKEDPQETGE FT PAGNTATTTAPAGDDAVRTEHLYKHPLMNVWTLWYLENDRSKSWEDMQNEITSFDTVED FT FWSLYNHIKPPSEIKLGSDYSLFKKNIRPMWEDAANKQGGRWVITLNKSSKTDLDNLWL FT DVLLCLIGEAFDHSDQICGAVINIRGKSNKISIWTADGNNEEAALEIGHKLRDALRLGR FT NNSLQYQLHKDTMVKQGSNVKSIYTL"
Following the mRNA feature is the CDS feature shown above, describing the region that ultimately encodes the protein product. Focusing just on eukaryotic initiation factor 4E-II, the CDS feature also shows a joinline with coordinates that are slightly different from those shown in the mRNA feature, specifically at the beginning and end positions. The difference lies in the fact that the 5′ and 3′ untranslated regions (UTRs) are included in the mRNA feature but not in the CDS feature. The CDS feature corresponds to the sequence of amino acids found in the translated protein product whose sequence is shown in the /translationqualifier above. The /codon_startqualifier indicates that the amino acid translation of the first codon begins at the first position of this joined region, with no offset.
The /protein_idqualifier shows the accession number for the corresponding entry in the protein databases (AAC03524.1) and is hyperlinked, enabling the user to go directly to that entry. These unique identifiers use a “3 + 5” format – three letters, followed by five numbers. Versions are indicated by the decimal that follows; when the protein sequence in the record changes, the version is incremented by one. The assignment of a gene product or protein name (via the /proteinqualifier) often is subjective, sometimes being assigned via weak similarities to other (and sometimes poorly annotated) sequences. Given the potential for the transitive propagation of poor annotations (that is, bad data tend to beget more bad data), users are advised to consult curated nucleotide and protein sequence databases for the most up-to-date, accurate information regarding the putative function of a given sequence. Finally, notice the extensive cross-referencing via the /db_xrefqualifier to entries in InterPro, the Protein Data Bank (PDB), and UniProtKB/Swiss-Prot, as well as to a Gene Ontology annotation (GOA; Gene Ontology Consortium 2017).
Implicit in the source feature and the organism that is assigned to it is the genetic code used to translate the nucleic acid sequence into a protein sequence when a CDS feature is present in the record. Also, the DNA-centric nature of these feature tables means that all features are mapped through a DNA coordinate system, not that of amino acid reference points, as shown in the examples in Appendices 1.3and 1.4.
SQ Sequence 2881 BP; 849 A; 699 C; 585 G; 748 T; 0 other; cggttgcttg ggttttataa catcagtcag tgacaggcat ttccagagtt gccctgttca 60 acaatcgata gctgcctttg gccaccaaaa tcccaaactt aattaaagaa ttaaataatt 120 cgaataataa ttaagcccag taacctacgc agcttgagtg cgtaaccgat atctagtata 180 . . . aaacggaacc ccctttgtta tcaaaaatcg gcataatata aaatctatcc gctttttgta 2820 gtcactgtca ataatggatt agacggaaaa gtatattaat aaaaacctac attaaaaccg 2880 g 2881 //
Finally, at the end of every nucleotide sequence record, one finds the actual nucleotide sequence, with 60 bases per row. Note that, in the SQ line signaling the beginning of this section of the record, not only is the overall length of the sequence provided, but a count of how many of each individual type of nucleotide base is also provided, making it quite easy to compute the GC content of this sequence.
Graphical interfaces have been developed to facilitate the interpretation of the data found within text-based flatfiles, with an example of the graphical view of the ENA record for our sequence of interest (U54469.1) shown in Figure 1.1. These graphical views are particularly useful when there is a long list of documented biological features within the feature table, enabling the user to visualize potential interactions or relationships between biological features. An additional example of the use of graphical views to assist in the interpretation of the information found within a database record is provided in the discussion of the NCBI Entrez discovery pathway in Chapter 2, as well as later in this chapter.
As one might expect, especially given the breakneck speed at which DNA sequence data are currently being produced, there is a significant amount of redundancy within the major sequence databases, with a good number of sequences being represented more than once. This is often problematic for the end user, who may find themselves confused as to which sequence to use after performing a search that returns numerous results. To address this issue, NCBI developed RefSeq, the goal of which is to provide a single reference sequence for each molecule of the central dogma – DNA, RNA, and protein. The distinguishing features of RefSeq go beyond its non-redundant nature, with individual entries including the biological attributes of the gene, gene transcript, or protein. RefSeq entries encompass a wide taxonomic range, and entries are updated and curated on an ongoing basis to reflect current knowledge about the individual entries. Additional information on RefSeq can be found in Box 1.2.
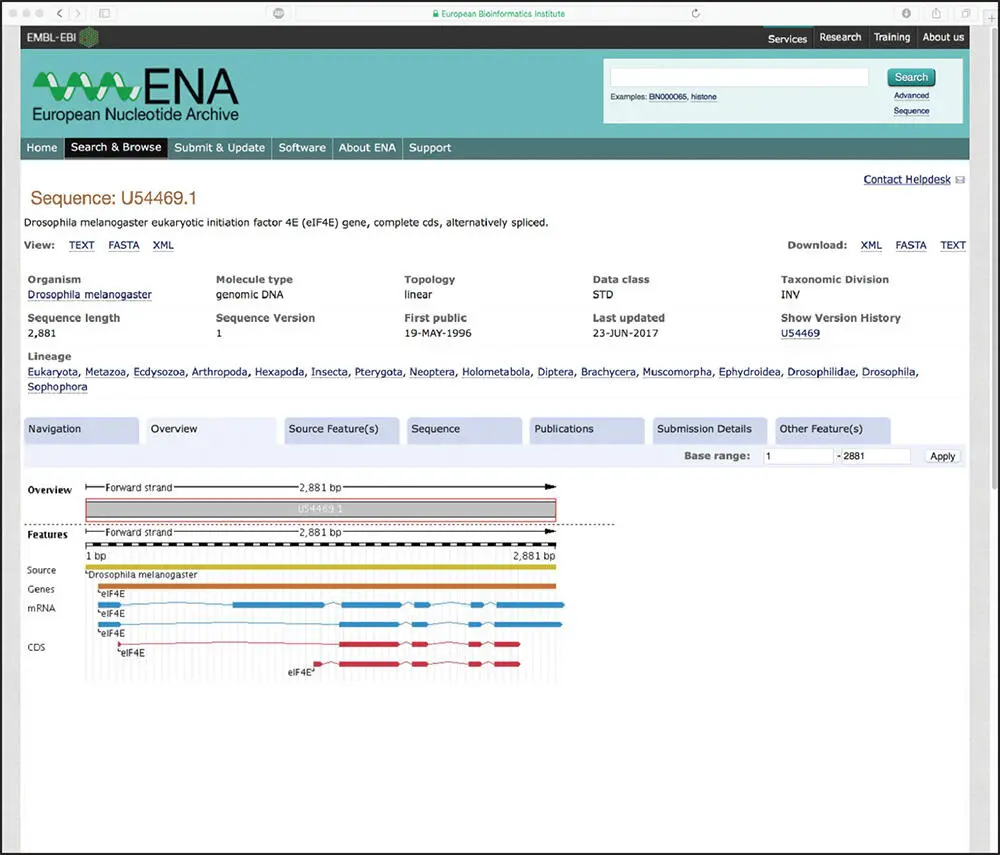
Figure 1.1 The landing page for ENA record U54469.1, providing a graphical view of biological features found within the sequence of the Drosophila melanogaster eukaryotic initiation factor 4E ( eIF4E ) gene. The tracks within the graphical view show the position of the gene, mRNAs, and coding regions (marked CDS) within the 2881 bp sequence reported in this record.
The first several chapters of this book describe a variety of ways in which sequence data and sequence annotations find their way into public databases. While the combination of data derived from systematic sequencing projects and individual investigators' laboratories yields a rich and highly valuable set of sequence data, some problems are apparent. The most important issue is that a single biological entity may be represented by many different entries in various databases. It also may not be clear whether a given sequence has been experimentally determined or is simply the result of a computational prediction.
To address these issues, NCBI developed the RefSeq project, the major goal of which is to provide a reference sequence for each molecule in the central dogma (DNA, mRNA, and protein). As each biological entity is represented only once, RefSeq is, by definition, non-redundant. Nucleotide and protein sequences in RefSeq are explicitly linked to one another. Most importantly, RefSeq entries undergo ongoing curation, assuring that the RefSeq entry represents the most up-to-date state of knowledge regarding a particular DNA, mRNA, or protein sequence.
RefSeq entries are distinguished from other entries in GenBank through the use of a distinct accession number series. RefSeq accession numbers follow a “2 + 6” format: a two-letter code indicating the type of reference sequence, followed by an underscore and a six-digit number. Experimentally determined sequence data are denoted as follows:
Читать дальше





