NT_123456 |
Genomic contigs (DNA) |
NM_123456 |
mRNAs |
NP_123456 |
Proteins |
Reference sequences derived through genome annotation efforts are denoted as follows:
XM_123456 |
Model mRNAs |
XM_123456 |
Model proteins |
It is important to understand the distinction between the “N” numbers and “X” numbers – the former represent actual, experimentally determined sequences, while the latter represent computational predictions derived from the raw DNA sequence.
Additional types of RefSeq entries, along with more information on the RefSeq project, can be found on the NCBI RefSeq web site.
Protein Sequence Databases
With the availability of myriad complete genome sequences from both prokaryotes and eukaryotes, significant effort is being dedicated to the identification and functional analysis of the proteins encoded by these genomes. The large-scale analysis of these proteins continues to generate huge amounts of data, including through the use of proteomic methods ( Chapter 11) and through protein structure analysis ( Chapter 12), to name a few. These and other methods make it possible to identify large numbers of proteins quickly, to map their interactions ( Chapter 13), to determine their location within the cell, and to analyze their biological activities. This ever-increasing “information space” reinforces the central role that protein sequence databases play as a resource for storing data generated by these efforts, making them freely available to the life sciences community.
As most sequence data in protein databases are derived from the translation of nucleotide sequences, they can be, in large part, thought of as “secondary databases.” Universal protein sequence databases cover proteins from all species, whereas specialized protein sequence databases concentrate on particular protein families, groups of proteins, or those from a specific organism. Representative model organism databases include the Mouse Genome Database (MGD; Smith et al. 2018) and WormBase (Lee et al. 2018), among others (Baxevanis and Bateman 2015; Rigden and Fernández 2018). Organismal sequence databases are discussed in greater detail in Chapter 2.
Universal protein databases can be divided further into two broad categories: sequence repositories, where the data are stored with little or no manual intervention, and curated databases, in which experts enhance the original data through expert biocuration . The importance of ensuring interoperability, creating and implementing standards, and adopting best practices aimed at accurately representing the biological knowledge found within the sequence databases is absolutely paramount. Indeed, these curation goals are so important that there is an organization called the International Society for Biocuration, the primary mission of which is to advance these central tenets.
The NCBI Protein Database
NCBI maintains the Protein database, which derives its content from a number of sources. These include the translations of the annotated coding regions from INSDC databases described above, from RefSeq ( Box 1.2), and from NCBI's Third Party Annotation (TPA) database. The TPA dataset is quite interesting in its own right, as it captures both experimental and inferential data provided by the scientific community to supplement the information found in an INSDC nucleotide entry. As the name suggests, the information in the TPA is provided by third parties and not by the original submitter of the corresponding INSDC entry. The NCBI Protein database also includes additional non-NCBI sources of protein sequence data, including Swiss-Prot, PIR, PDB, and the Protein Research Foundation. Step-by-step methods for performing searches against the NCBI Protein database are described in detail in Chapter 3.
Although data repositories are an essential vehicle through which scientists can access sequence data as quickly as possible, it is clear that the addition of biological information from multiple, highly regarded sources greatly increases the power of the underlying sequence data. The UniProt Consortium was formed to accomplish just that, bringing together the Swiss-Prot, TrEMBL, and the Protein Information Resource Protein Sequence Database under a single umbrella, called UniProt (UniProt Consortium 2017). UniProt comprises three main databases: the UniProt Archive, a non-redundant set of all publicly available protein sequences compiled from a variety of source databases; UniProtKB, combining entries from UniProtKB/Swiss-Prot and UniProtKB/TrEMBL; and the UniProt Reference Clusters (UniRef), containing non-redundant views of the data contained in UniParc and UniProtKB that are clustered at three different levels of sequence identity (Suzek et al. 2015).
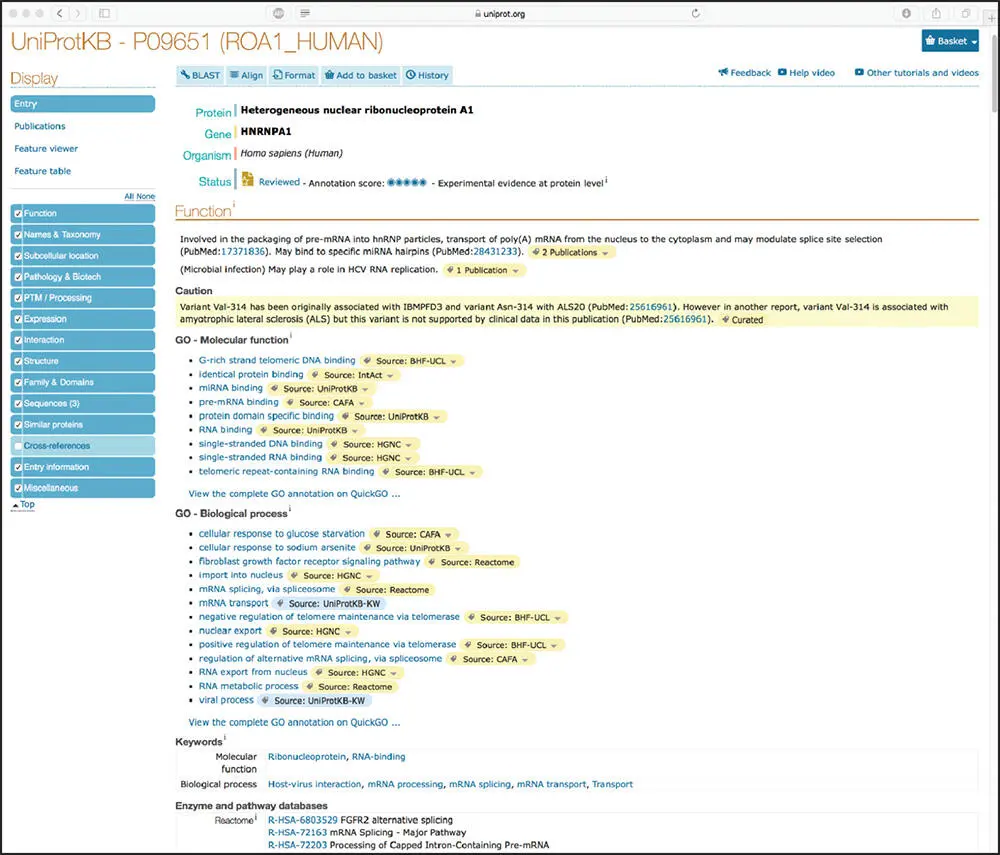
Figure 1.2 Results of a search for the human heterogeneous nuclear ribosomal protein A1 record within UniProtKB, using the accession number P09651 as the search term. See text for details.
The wealth of information found within a UniProtKB entry can be best illustrated by an example. Here, we will consider the entry for the human heterogeneous nuclear ribonuclear protein A1, with accession number P09651. A search of UniProtKB using this accession number as the search term produces the view seen in Figure 1.2. The lower part of the left-hand column shows the various types of information available for this protein, and the user can select or de-select sections based on their interests. The main part of the window provides basic identifying information about this sequence, as well as an indication of whether the entry has been manually reviewed and annotated by UniProtKB curators. Here, we see that the entry has indeed been reviewed and that there is experimental evidence that supports the existence of the protein. The next section in the file is devoted to conveying functional information, also providing Gene Ontology (GO) terms that are associated with the entry, as well as links to enzyme and pathway databases such as Reactome (see Chapter 13). Clicking on any of the blue tiles in the left-hand column will jump the user down to the selected section of the entry. For instance, if one clicks on Subcellular location, the view seen in Figure 1.3is produced, providing a color-coded schematic of the cell indicating the type of annotation (manual or automatic) and links to publications supporting the annotation. The lower part of Figure 1.3also shows information regarding the protein's involvement in disease, documenting variants that have been implicated in early onset Paget disease and amyotrophic lateral sclerosis (Kim et al. 2013; Liu et al. 2016).

Figure 1.3 The Subcellular location and Pathology & Biotech sections of the record for the human heterogeneous nuclear ribosomal protein A1 record within UniProtKB. These sections can be accessed by clicking on the blue tiles in the left-hand column of the window. See text for details.
In the upper left corner of the UniProtKB window are display options that are quite useful in visualizing the significant amount of data found in this entry's feature table. By clicking on Feature viewer, one is presented with the view shown in Figure 1.4, neatly summarizing the annotations for this sequence in a coordinate-based fashion. Any of the sections can be expanded by clicking on the labels in the blue boxes to the left of the graphic. Here, the post-translational modification (PTM) section has been expanded, showing the position of modified residues in this protein; clicking on any of the markers in the track will produce a pop-up with additional information on the PTM, along with relevant links to the literature. In Figure 1.5, the Structural features and Variants sections have also been expanded, showing the positions of all alpha helices, beta strands, and beta turns within the protein, as well as the location of putatively clinically relevant point mutations. Here, a variant at position 351 is highlighted, with the proline-to-leucine variant identified as part of the ClinVar project (Landrum et al. 2016) having a possible association with relapsing–remitting multiple sclerosis. By examining different sections of this very useful graphical display, the user can start to see how various features overlap with one another, perhaps indicating whether a known or predicted disease-causing variant falls within a structured region of the protein. These annotations and observations can provide important insights with respect to experimental design and the interpretation of experimental data.
Читать дальше






