By using approaches such as VAST and VAST+, it is possible to find structural relationships between proteins in cases where simply looking at sequence similarity may not suggest relatedness – information that could, with additional data and insights, be used to help inform the question of functional similarity. More information on additional structure prediction methods based on X-ray or nuclear magnetic resonance (NMR) coordinate data can be found in Chapter 12.
Weighted Key TermsThe problem of comparing sequence or structure data somewhat pales next to that of comparing PubMed entries, which consist of free text whose rules of syntax are not necessarily fixed. Given that no two people's writing styles are exactly the same, finding a way to compare seemingly disparate blocks of text poses a substantial problem. Entrez employs a method known as the relevance pairs model of retrieval to make such comparisons, relying on weighted key terms (Wilbur and Coffee 1994; Wilbur and Yang 1996). This concept is best described by example. Consider two manuscripts with the following titles:
BRCA1 as a Genetic Marker for Breast Cancer
Genetic Factors in the Familial Transmission of the Breast Cancer BRCA1 Gene
Both titles contain the terms BRCA1 , Breast , and Cancer , and the presence of these common terms may indicate that the manuscripts are similar in subject matter. The proximity between the words is also considered, so that words common to two records that are closer together are scored higher than common words that are further apart. In the example, the terms Breast and Cancer are always next to each other, so they would score higher based on proximity than either of those words would against BRCA1 . Common words found in a title score higher than those found in an abstract, since title words are presumed to be “more important” than those found in the body of an abstract. Overall, weighting depends inversely on the frequency of a given word among all the entries in PubMed, with words that occur infrequently in the database assigned a higher weight while common words are down-weighted.
The hard link concept is simpler and much more straightforward than the neighboring approaches described above. Hard links are applied between entries in different databases and exist wherever there is a logical connection between entries. For instance, if a PubMed entry describes the sequencing of a chromosomal region containing a gene of interest, a hard link is established between the PubMed entry and the corresponding nucleotide entry for that gene. If an open reading frame in that gene codes for a known protein, a hard link is established between the nucleotide entry and the protein entry. If the protein entry has an experimentally deduced structure, a hard link would be placed between the protein entry and the structural entry.
Searches can begin anywhere within the Entrez ecosystem – there are no constraints on the user as to where the foray into this information space must begin. However, depending on which database is used as the jumping-off point, different database fields will be available for searching. This stands to reason, as the entries in different databases are necessarily organized differently, reflecting the biological nature of the entities that each database is trying to catalog.
The Entrez Discovery Pathway
The best way to illustrate the integrated nature of the Entrez system and to drive home the power of neighboring is by considering some biological examples. The simplest way to query Entrez is through the use of individual search terms, coupled together by Boolean operators such as AND, OR, or NOT. Consider the case in which one wants to retrieve all available information on a gene named DCC (deleted in colorectal carcinoma), limiting the returned information to publications where an investigator named Guy A. Rouleau is an author. There is a very simple query interface at the top of the NCBI home page, allowing the user to select which database they want to search from a pull-down menu and a text box where the query terms can be entered. In this case, to search for published papers, PubMed would be selected from the pull-down menu and, within the text box to the right, the user would type DCC AND "Rouleau GA" [AU]. The [AU]qualifying the second search term indicates to Entrez that this is an author term, so only the author field in entries should be considered when evaluating this part of the search statement. The result of the query is shown in Figure 2.2. Here, three entries matching the query were found in PubMed. The user can further narrow down the query by adding additional terms if the user is interested in a more specific aspect of this gene or if there are quite simply too many entries returned by the initial query. A list of available field delimiters is given in Table 2.1.
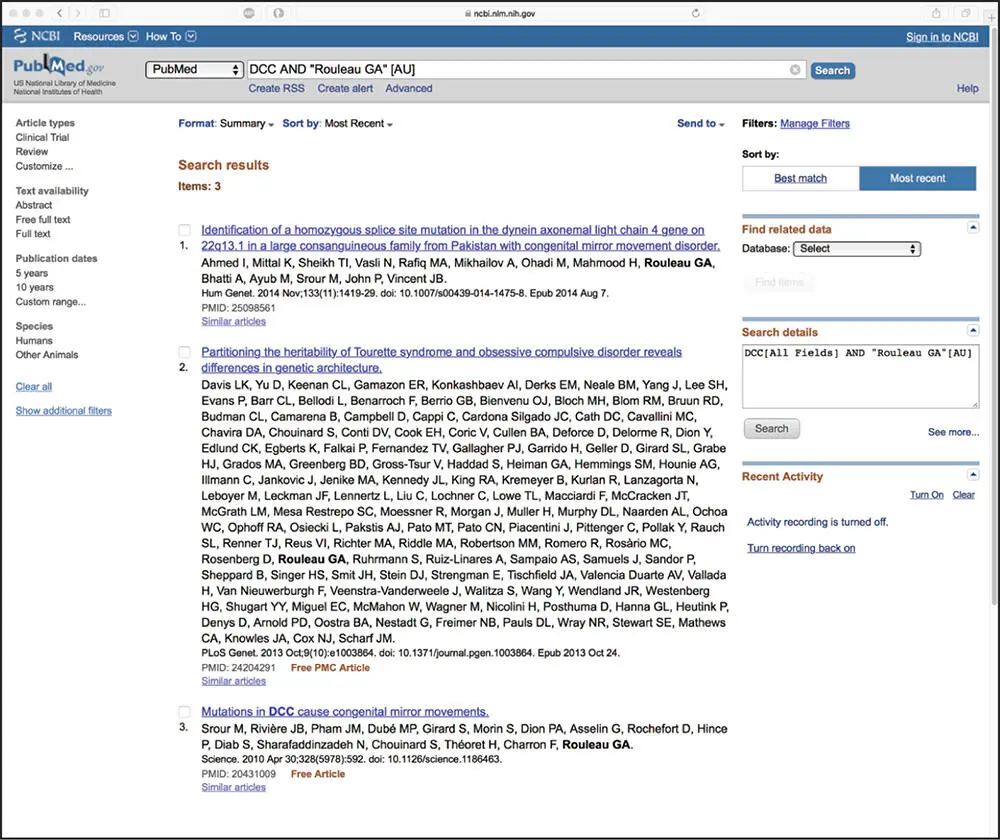
Figure 2.2 Results of a text-based Entrez query against PubMed using Boolean operators and field delimiters. The initial query ( DCC AND "Rouleau GA" [AU]) is shown in the search box near the top of the window, with the three papers identified using this query following below. Each entry gives the title of the manuscript, the names of the authors, and the citation information. The actual record can be retrieved by clicking on the name of the manuscript.
Table 2.1 Entrez Boolean search statements.
General syntax: search term [tag] Boolean operator search term [tag] ...where [tag]= |
[ACCN] |
Accession |
[AD] |
Affiliation |
[ALL] |
All fields |
[AU] |
Author name Lentz R [AU] yields all of Lentz RA, Lentz RB, etc. "Lentz R" [AU] yields only Lentz R |
[AUID] |
Unique author identifier, such as an ORCID ID |
[ECNO] |
Enzyme Commission numbers |
[EDAT] |
Entrez date YYYY/MM/DD, YYYY/MM, or YYYY; insert a colon for date range, e.g. 2016:2018 |
[GENE] |
Gene name |
[ISS] |
Issue of journal |
[JOUR] |
Journal title, official abbreviation, or ISSN number Journal of Biological Chemistry J Biol Chem 0021-9258 |
[LA] |
Language |
[MAJR] |
MeSH major topic One of the major topics discussed in the article |
[MH] |
MeSH terms Controlled vocabulary of biomedical terms (subject) |
[ORGN] |
Organism |
[PDAT] |
Publication date YYYY/MM/DD, YYYY/MM, or YYYY; insert a colon for date range, e.g. 2016:2018 |
[PMID] |
PubMed ID |
[PROT] |
Protein name (for sequence records) |
[PT] |
Publication type, includes: Review Clinical Trial Lectures Letter Technical Report |
[SH] |
MeSH subheading Used to modify MeSH Terms stenosis [MH] AND pharmacology [SH] |
[SUBS] |
Substance name Name of chemical discussed in article |
[SI] |
Secondary source ID Names of secondary source databanks and/or accession numbers of sequences discussed in article |
[TITL] |
Title word Only words in the definition line (not available in Structure database) |
[WORD] |
Text words All words and numbers in the title and abstract, MeSH terms, subheadings, chemical substance names, personal name as subject, and MEDLINE secondary sources |
[VOL] |
Volume of journal |
and Boolean operator = AND, OR, or NOT |
For each of the found papers shown in the Summary view in Figure 2.2, the user is presented with the title of the paper, the authors of that paper, and the citation. To look at any of the papers resulting from the search, the user can simply click on any of the hyperlinked titles. For this example, consider the third reference in the list, by Srour et al. (2010). Clicking on the title takes the user to the Abstract view shown in Figure 2.3. This view presents the name of the paper, the list of authors, their institutional affiliation, and the abstract itself. Below the abstract is a gray bar labeled “MeSH terms, Substances”; clicking on the plus sign at the end of the gray bar reveals cataloging information (MeSH terms, for me dical s ubject h eadings) and indexed substances related to the manuscript. Several alternative formats are available for displaying this information, and these various formats can be selected using the Format pull-down menu found in the upper left corner of the window. Switching to MEDLINE format produces the MEDLINE layout, with two-letter codes corresponding to the contents of each field going down the left-hand side of the entry (e.g. the author field is again denoted by the code AU). Lists of entries in this format can be saved to the desktop and easily imported into third-party bibliography management programs.
Читать дальше





