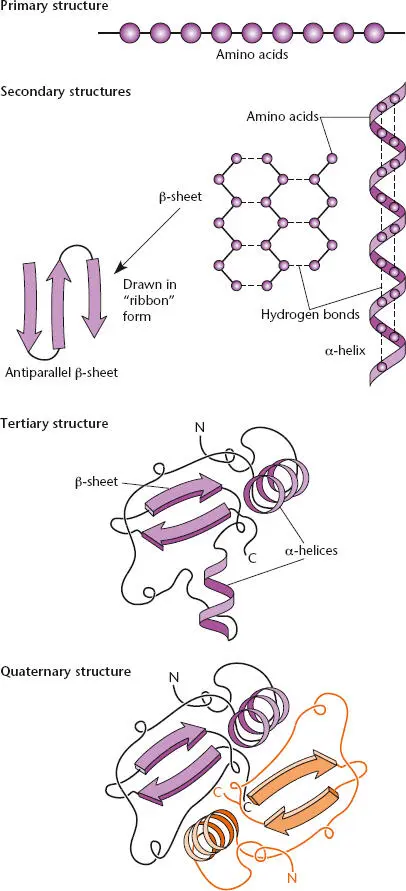
Figure 2.20 Primary, secondary, tertiary, and quaternary structures of proteins.
Polypeptides usually also have a well-defined tertiary structure, in which they fold up on themselves with hydrophobic amino acids (such as leucine and isoleucine), which are not very soluble in water, on the inside and charged amino acids (such as glutamate and lysine), which are more water soluble, or hydrophilic, on the outside. We discuss the structure of proteins in more detail in “Protein Folding and Degradation” below.
Proteins made up of more than one polypeptide chain also have quaternary structure. Such proteins are called multimeric proteins. When the polypeptides are the same, the protein is a homomultimer. When they are different, the protein is a heteromultimer. Other names reflect the number of polypeptides in the protein. For example, the term homodimerdescribes a protein made of two identical polypeptides, whereas heterodimerdescribes a protein made of two different polypeptides. The names trimer, tetramer, and so on refer to increasing numbers of polypeptides. Hence, the ρ transcription termination factor is a homohexamer (see above).
The polypeptide chains in a protein are usually held together by hydrogen bonds. The only covalent chemical bonds in most proteins are the peptide bonds that link adjacent amino acids to form the polypeptide chains. As a result, if a multimeric protein is heated, it falls apart into its individual polypeptide chains. However, some proteins are unusually stable; these include extracellular enzymes, which must be able to function in the harsh environment outside the cell. Such proteins are often also held together by disulfide bondsbetween cysteine amino acids in the protein.
The translation of the sequence of nucleotides in mRNA into the sequence of amino acids in a protein occurs on the ribosome. The overall process of translation is highly conserved in bacteria, archaea, and eukaryotes, and the machinery is also highly conserved. As mentioned in “rRNAs and tRNAs” above, the ribosome is one of the largest and most complicated structures in cells, consisting of three different RNAs and over 50 different proteins in bacteria. It is also one of the major constituents of the bacterial cell, and much of the biosynthetic capacity of the cell goes into making ribosomes. Each cell contains thousands of ribosomes, with the actual number depending on the growth conditions. It is also one of the most evolutionarily highly conserved structures in cells, having remained largely unchanged in shape and structure from bacteria to humans. The key role of protein synthesis in the cell has led to the development of important antibiotics that target the translational machinery ( Box 2.3).
The ribosome is an enormous enzyme that performs the complicated role of polymerizing amino acids into polypeptide chains, using the information in mRNA as a guide. As such, a better name for it might have been protein polymerase, by analogy to DNA and RNA polymerases. The historical name “ribosome” was coined before its function was known, because it is large enough to have been visualized under the electron microscope and so it was called a “some” (for body) and “ribosome” because it contains ribonucleotides. The recent determination of the structure of the ribosome (see below) has led to important insights into how it performs its function of polymerizing amino acids.
Structure of the Bacterial Ribosome
Figure 2.21shows the components of a bacterial ribosome. The complete ribosome, called the 70S ribosome in bacteria, consists of two subunits, the 30S subunit, which contains one molecule of 16S rRNA, and the 50S subunit, which contains one molecule each of 23S and 5S rRNA. Each subunit also contains ribosomal proteins; the 30S subunit contains approximately 21 different proteins (S1, S2, etc., where S indicates small subunit proteins), while the 50S subunit contains approximately 31 different proteins (L1, L2, etc., where L indicates large subunit proteins). Ribosomes from different bacterial species have very similar compositions, with small differences in the number of ribosomal proteins. Like the names for the different types of rRNA, the names of ribosomal subunits are derived from their sedimentation rates during ultracentrifugation. The 30S and 50S subunits normally exist separately in the cell and come together to form the complete 70S ribosome only when they are translating an mRNA. Note that sedimentation values are not additive; the complete ribosome is 70S in size, despite being composed of subunits that are 30S and 50S in size. This is because the two subunits fit together into a tight complex.
The two ribosomal subunits play very different roles in translation. To initiate translation, the 30S subunit first binds to the mRNA. Then, the 50S ribosome binds to the 30S subunit to make the 70S ribosome. From that point on, the 30S subunit mostly helps to select the correct aminoacyl-tRNA (aa-tRNA)for each codon while the 50S subunit does most of the work of forming the peptide bonds and translocating the tRNAs from one site on the ribosome to another (see below). The 70S ribosome moves along the mRNA, allowing tRNA anticodons to pair with the mRNA codons and translate the information from the nucleic acid chain into a polypeptide. After the polypeptide chain is completed, the ribosome separates again into the 30S and 50S subunits. The role of the subunits is discussed in more detail below.
BOX 2.3
Antibiotic Inhibitors of Translation
The translation apparatus of bacteria is a particularly tempting target for antibacterial drugs because it is somewhat different from the eukaryotic translation apparatus and is highly conserved among bacteria. Antibiotics that inhibit translation are among the most useful of all the antibiotics and are listed in the table, which also lists their targets and sources. Toxicity in some cases may result from the similarity of bacterial and mitochondrial ribosomes. Some of these antibiotics are also very useful in combating fungal diseases and in cancer chemotherapy.
Antibiotics that block translation
| Antibiotic |
Source |
Target |
| Puromycin |
Streptomyces alboniger |
Ribosomal A site |
| Kanamycin |
Streptomyces kanamyceticus |
16S rRNA |
| Neomycin |
Streptomyces fradiae |
16S rRNA |
| Streptomycin |
Streptomyces griseus |
30S ribosome |
| Thiostrepton |
Streptomyces azureus |
23S rRNA |
| Gentamicin |
Micromonospora purpurea |
16S rRNA |
| Tetracycline |
Streptomyces rimosus |
Ribosomal A site |
| Chloramphenicol |
Streptomyces venezuelae |
Peptidyltransferase |
| Erythromycin |
Saccharopolyspora erythraea |
23S rRNA |
| Fusidic acid |
Fusidium coccineum |
EF-G |
| Kirromycin |
Streptomyces collinus |
EF-Tu |
Inhibitors that Mimic tRNA
Puromycin mimics the 3′ end of tRNA with an amino acid attached (aa-tRNA). It enters the ribosome as does an aa-tRNA, and the peptidyltransferase attaches it to the growing polypeptide. However, it does not translocate properly from the A site to the P site, and the peptide with puromycin attached to its carboxyl terminus is released from the ribosome, terminating translation.
Читать дальше












