Before translation can begin, a specific amino acid is attached to each tRNA by its cognate aaRS( Figure 2.24). Each of these enzymes specifically recognizes only one amino acid and one class of tRNA—hence the name cognate . How each aaRS recognizes its own tRNA varies, but the anticodon (i.e., the three tRNA nucleotides that base pair with the complementary mRNA sequence [ Figure 2.25]) is not the only determinant. Other variations in the tRNA structure, such as the identity of the discriminator base immediately upstream of the CCA at the 3′ end ( Figure 2.17) and the size of the variable loop, also contribute to aaRS recognition specificity. In some cases, if the anticodon in a given tRNA is mutated, the cognate aaRS still attaches the amino acid for the original tRNA, and that amino acid is inserted at a different codon in the mRNA. This is the basis of nonsense suppression, which is discussed in chapter 3. The amino acid is attached to the terminal A residue on the tRNA, and the energy for the reaction is provided by cleavage of ATP. Finally, the tRNA with its amino acid is bound to the protein EF-Tu, which assists in delivery of the aa-tRNA to the ribosome.
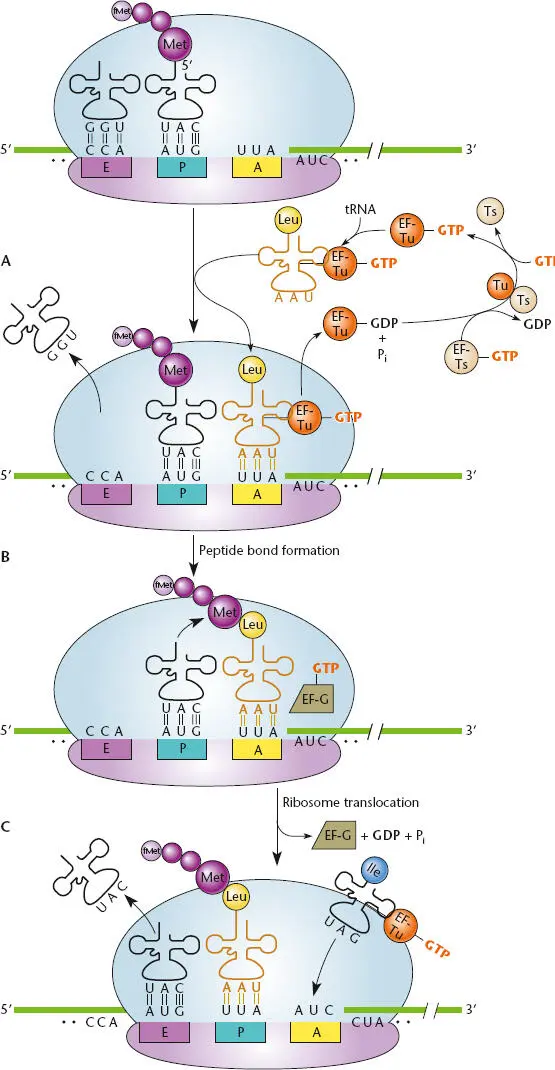
Figure 2.23 Overview of translation. (A)The ribosomal A (aminoacyl) site is empty, the growing peptide chain is attached to the P (peptidyl) site tRNA, and the tRNA that previously contained the peptide chain is in the E (exit) site. (B)The tRNA bound to its amino acid and complexed with elongation factor Tu (EF-Tu) and guanosine triphosphate (GTP) comes into the empty A site and remains there if its anticodon matches the mRNA codon at that site; the E site tRNA leaves the ribosome. EF-Tu–GTP is converted to EF-Tu–GDP and released from the ribosome, then is recycled to EF-Tu–GTP by EF-Ts. (C)Peptidyltransferase (23S rRNA in the 50S ribosome) catalyzes peptide bond formation between the carboxyl end of the growing polypeptide carried by the P site tRNA and the amino end of the amino acid carried by the A site tRNA. (D)Translation elongation factor G (EF-G) catalyzes translocation of the A site tRNA to the P site, making room at the A site for another aminoacyl-tRNA. The previous P site tRNA, now stripped of its polypeptide, moves to the E site before exiting the ribosome.
Table 2.2The genetic code
| First position (5′ end) |
Second position |
Third position (3′ end) |
| U |
C |
A |
G |
| U |
Phe |
Ser |
Tyr |
Cys |
U |
|
Phe |
Ser |
Tyr |
Cys |
C |
|
Leu |
Ser |
Stop |
Stop |
A |
|
Leu |
Ser |
Stop |
Trp |
G |
| C |
Leu |
Pro |
His |
Arg |
U |
|
Leu |
Pro |
His |
Arg |
C |
|
Leu |
Pro |
Gln |
Arg |
A |
|
Leu |
Pro |
Gln |
Arg |
G |
| A |
Ile |
Thr |
Asn |
Ser |
U |
|
Ile |
Thr |
Asn |
Ser |
C |
|
Ile |
Thr |
Lys |
Arg |
A |
|
Met |
Thr |
Lys |
Arg |
G |
| G |
Val |
Ala |
Asp |
Gly |
U |
|
Val |
Ala |
Asp |
Gly |
C |
|
Val |
Ala |
Glu |
Gly |
A |
|
Val |
Ala |
Glu |
Gly |
G |
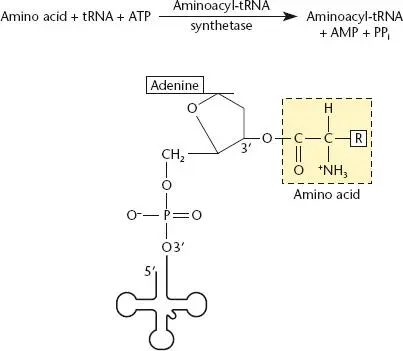
Figure 2.24 Aminoacylation of a tRNA by its cognate aminoacyl-tRNA synthetase (aaRS). ATP is used as a source of energy, and the amino acid is attached to the adenosine residue at the 3′ end of the tRNA. Each amino acid utilizes a dedicated aaRS.
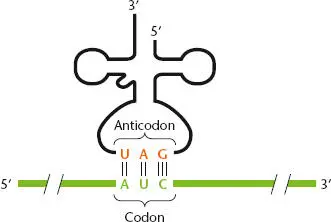
Figure 2.25 Complementary pairing between a tRNA anticodon and an mRNA codon. The codon (green) is shown 5′-3′, and the tRNA is shown 3′-5′ to allow antiparallel pairing between the codon and the tRNA anticodon (orange).
TRANSLATION INITIATION REGIONS
In the chain of thousands of nucleotides that make up an mRNA, the ribosome must bind and initiate translation at the correct site. If the ribosome starts working at the wrong position, the protein will have the wrong N-terminal amino acids and/or the mRNA will be translated out of frame and all of the amino acids will be wrong. Hence, mRNAs have sequences called translational initiation regions (TIRs)or ribosome-binding sites (RBSs)that flag the correct first codon for the ribosome. In spite of extensive research, it is still not possible to predict with 100% accuracy whether a sequence is a TIR. However, some general features of TIRs are known.
All bacterial and archaeal TIRs have an initiation codon, which is recognized by a dedicated initiator tRNA. This tRNA is always aminoacylated with methionine by methionyl-tRNA synthetase, and the methionine is further modified by addition of a formyl group ( Figure 2.26). The three bases in initiator codons are usually AUG or GUG but in rare cases are UUG or CUG. Regardless of which amino acid these sequences call for in the genetic code (Table 2.2), they encode methionine (actually formylmethionine) as the N-terminal amino acid if they are serving as initiation codons. After translation, this methionine is often cut off by an aminopeptidase (see below). Notice that for the initiation codons, the first position of the codon can mispair with the tRNA anticodon, which always matches AUG; this differs from “wobble” during translation elongation, which involves mismatches at the third position of the codon (see below).
The initiation codon does not have to be the first sequence in the mRNA chain. In fact, the 5′ end of the mRNA may be some distance from the initiation codon of the first coding region in a transcript; this intervening region is called the 5′ untranslated region(5′-UTR) or leader region. In bacteria, many transcripts contain multiple coding regions. The translation initiation complex binds internally to the mRNA to identify the TIR for each coding region. This is different from translation in eukaryotes, where the translation initiation complex usually binds to the 5′ end of the transcript and initiates translation at the first AUG codon it encounters that is accessible (see below).
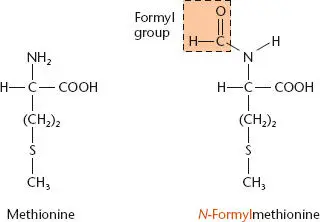
Figure 2.26 Conversion of methionine (Met) to N -formyl-methionine (fMet) by transformylase.
Читать дальше















