A variety of physical techniques, combined with much indirect information accumulated over the years from genetics and biochemistry, have revealed many details of the overall structure of the ribosome. The crystal structures of the individual subunits and the entire 70S ribosome have been determined and correlated with the earlier indirect information. Many laboratories participated in this project, and this awesome achievement will go down in history as one of the major milestones in molecular biology, recognized by the Nobel Prize. We can review only a few of the most salient features here.
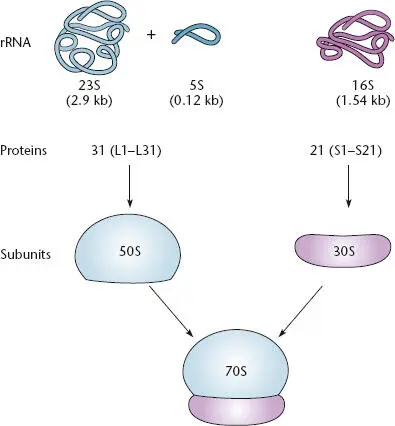
Figure 2.21 The composition of a bacterial ribosome containing one copy each of the 16S, 23S, and 5S rRNAs, as well as many proteins. The proteins of the large 50S subunit are designated L1 to L31. The proteins of the small 30S subunit are designated S1 to S21. The 30S and 50S subunits combine to form the 70S ribosome, which carries out protein synthesis.
The two subunits of the ribosome are frequently represented as ovals, with a flat side that binds to the other subunit, leaving a small gap between them. It is through this gap that the mRNA moves, and the aa-tRNAs enter, interact with the mRNA, and pass through the ribosome, contributing their amino acids to the growing polypeptide chain. The newly synthesized polypeptide chain exits through a channel running through the 50S subunit. This channel is long enough to hold a chain of about 70 amino acids, so a polypeptide of this length must be synthesized before the N-terminal end of a protein first emerges from the ribosome.
The rRNAs play many of the most important roles in the ribosome, and the ribosomal proteins seem to be present mostly to give rigidity to the structure, helping to hold the rRNAs in place. This has contributed to speculation that RNAs were the primordial enzymes and that proteins came along later in the earliest stages of life on
Earth. The 23S rRNA, rather than a ribosomal protein, acts as the peptidyltransferase enzyme that performs the enzymatic function that forms the peptide bonds between the carboxyl end of the growing polypeptide and the amino group of the incoming amino acid. Thus, 23S rRNA is an RNA enzyme, or ribozyme. The 23S rRNA also forms most of the channel in the 50S subunit through which the growing polypeptide passes. The 16S RNA plays crucial roles in translation initiation and in matching each incoming aa-tRNA with the mRNA. A structure of the ribosome is shown in Figure 2.22.
The information in mRNA is interpreted by the pairing of consecutive triplets of nucleotides (codons) in the mRNA with the complementary anticodon sequences of the corresponding tRNAs. This pairing takes place on the ribosome, and accuracy of translation also depends on matching of a specific tRNA (with the appropriate anticodon) to the corresponding amino acid by a set of enzymes called aminoacyl-tRNA synthetase (aaRSs). aa-tRNAs generated by the aaRS enzymes are delivered to the ribosome by a protein factor called elongation factor-Tu (EF-Tu). During translation, the ribosome moves along the mRNA, and tRNAs interact with the mRNA at three distinct sites, designated the A (aminoacyl) site, the P (peptidyl) site, and the E (exit) site ( Figure 2.23). Each aa-tRNA is brought into the A site first, where its anticodon is tested for complementarity to the mRNA codon present at that site. If the anticodon is complementary to the codon, the tRNA is retained; if the anticodon does not match the codon, the tRNA is rejected, and a new aa-tRNA is brought into the A site. A match between the anticodon and the codon in the A site triggers the tRNA in the A site to interact with the tRNA already present in the P site. The P site tRNA carries the growing polypeptide chain, and the next step is transfer of the growing peptide chain from the P site tRNA to the A site tRNA. The amino acid carried on the A site tRNA is attached to the carboxyl end of the polypeptide, and the polypeptide (now 1 amino acid longer) is now linked to the A site tRNA. The now unattached P site tRNA moves to the E site of the ribosome and the A site tRNA (which now carries the polypeptide) moves to the P site of the ribosome, which results in an empty A site. Each tRNA retains contact with the mRNA through anticodon-codon pairing so that the mRNA moves through the ribosome in concert with the tRNAs. This results in placement of the next codon of the mRNA in the empty A site, which is now available for entry of another aa-tRNA. Movement of the unattached P site tRNA into the E site results in ejection of the previous unattached tRNA from the E site, which allows the next tRNA to enter the cycle. The details of this process are described below.
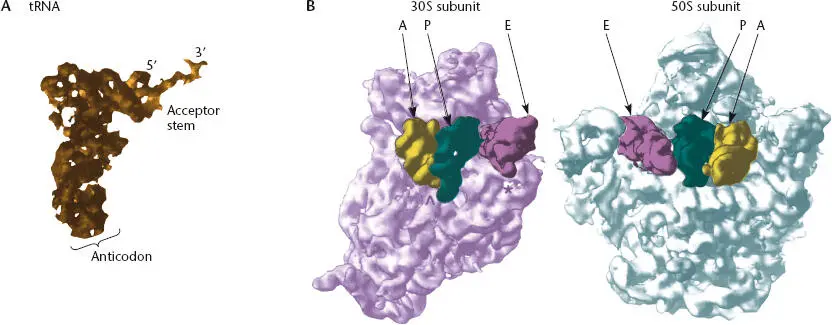
Figure 2.22 Crystal structures of a tRNA and the ribosome. (A)Structure of a tRNA. The anticodon loop is at the bottom, and the 3′ acceptor end, where the amino acid or growing polypeptide is attached, is at the top. (From Yusupov MM, Yusupova GZ, Baucom A, et al, Science 292:883–896, 2001. Modified with permission from AAaS.) (B)The two subunits of the ribosome separated and rotated to show the channel between them through which the tRNAs move. The 30S subunit is on the left, and the 50S subunit is on the right. The tRNAs bound at the A, P, and E sites are indicated in yellow, green, and purple, respectively. (From Cate JH, Yusupov MM, Yusupova G, et al, Science 285:209 5 –2014, 1999. Modified with permission from AAAS.)
Details of Protein Synthesis
In this section, we discuss the process of translation in more detail. First, we discuss reading frames, which determine which nucleotides in an mRNA are recognized as codons, and tRNA aminoacylation, which is responsible for correct matching of a tRNA with its cognate amino acid. Then, we discuss translation initiation, or how the 30S subunit finds the starting point of a coding sequence in the mRNA. Next, we discuss translation elongation, or what happens as the 70S ribosome moves along the mRNA, translating the information in the mRNA in the form of nucleotides into amino acids. Finally, we discuss how translation is terminated.
Each 3-nucleotide sequence, or codon, in the mRNA encodes a specific amino acid, and the assignment of the codons is known as the genetic code (Table 2.2). Because there are 3 nucleotides in each codon, an mRNA can be translated in three different frames in each region. Initiation of translation at a specific initiation codonestablishes the reading frame of translation, so that in most cases only a single reading frame is utilized. Once translation has begun, the ribosome moves 3 nucleotides at a time through the coding part of the mRNA. If the translation is occurring in the proper frame for protein synthesis, we say the translation is in the zero framefor that protein. If translation is occurring in the wrong reading frame, it can be displaced either back by 1 nucleotide in each codon (the –1 frame) or forward by 1 nucleotide (the +1 frame). In a few instances, translational frameshifts that change the reading frame even after translation has initiated can occur. Frameshift mutations (see chapter 3) cause incorrect reading of an mRNA because of the insertion or deletion of nucleotides in the DNA sequence, which is then copied into mRNA. Translational frameshifting occurs when the ribosome shifts its position on the mRNA without a change in the mRNA sequence itself.
Читать дальше













