MODULATION OF RNase ACTIVITY
The susceptibility of an RNA to different RNases can be affected by structural features of the RNA. RNA 3′ ends generated by termination of transcription at a factorindependent terminator contain an RNA hairpin, which inhibits binding of 3′-5′ exoribonucleases ( Figure 2.18). Degradation of RNAs of this type is often initiated by endonucleolytic cleavage, which removes the 3′ end of the RNA and allows the 5′ region of the molecule to be degraded. Degradation of the 3′ fragment can be initiated by polyadenylation of the 3′ end of the RNA by polyadenylate [poly(A)] polymerase, encoded by the pcnB gene. Addition of the poly(A) tail provides a “landing zone” for 3′-5′ exoribonucleases, which can initiate degradation of the poly(A) sequence and then continue to move through the terminator hairpin. This may be facilitated by colocalization of poly(A) polymerase and polynucleotide phosphorylase (PNPase; Table 2.1), one of the major 3′-5′ exonucleases, with other RNases into a complex called the degradosome. Note that polyadenylation of an mRNA in eukaryotes generally results in stabilization of the mRNA, while polyadenylation of an RNA in bacteria results in rapid degradation. Degradation of the 3′ fragment generated by endonucleolytic cleavage can also be directed by 5′-3′ exoribonucleases in organisms like B. subtilis that have this activity (see Condon, Suggested Reading). It is interesting to note that the 5′ ends of transcripts newly synthesized by RNA polymerase contain a triphosphate (from the initiating nucleotide), whereas the 5′ ends of RNAs generated by endonuclease cleavage contain monophosphates. The presence of a triphosphate protects the RNA, and this triphosphate can be removed by a dedicated enzyme, designated RppH, which enhances susceptibility to degradation by 5′-3′ exonucleases (see Hui et al., Suggested Reading).
Susceptibility to degradation can be used as a mechanism to regulate gene expression, because rapid degradation of an mRNA results in reduced synthesis of its protein product. Modulation of RNA stability can occur through changes in the RNA structure that affect RNase binding by binding of a regulatory protein to the RNA or by binding of a regulatory RNA. Mechanisms of this type are discussed in chapter 11.
Table 2.1Enzymes involved in mRNA processing and degradation
| Enzyme |
Substrate(s) |
Description |
| RNase E |
mRNA, rRNA, tRNA |
Endonuclease, highly conserved in all Proteobacteria and some Firmicutes (not B. subtilis ) |
| RNase III |
rRNA, polycistronic mRNA |
Endonuclease, cleaves double-stranded RNA in some stem-loops; found in most bacteria |
| RNase P |
Polycistronic mRNA, tRNA precursors |
Ribozyme, necessary to process 5′ ends of tRNAs |
| RNase G |
5′ end of 16S rRNA, mRNA |
Endonuclease, replaces RNase E in some bacteria |
| RNases J1 and J2 |
mRNA, rRNA |
5′-3′ exonuclease, endonuclease; found in most Firmicutes and some Proteobacteria bacteria (not E. coli ) |
| Poly(A) polymerase |
mRNA |
Found in most bacteria |
| PNPase |
mRNA, poly(A) tails |
3′-5′ exonuclease, found in all bacteria |
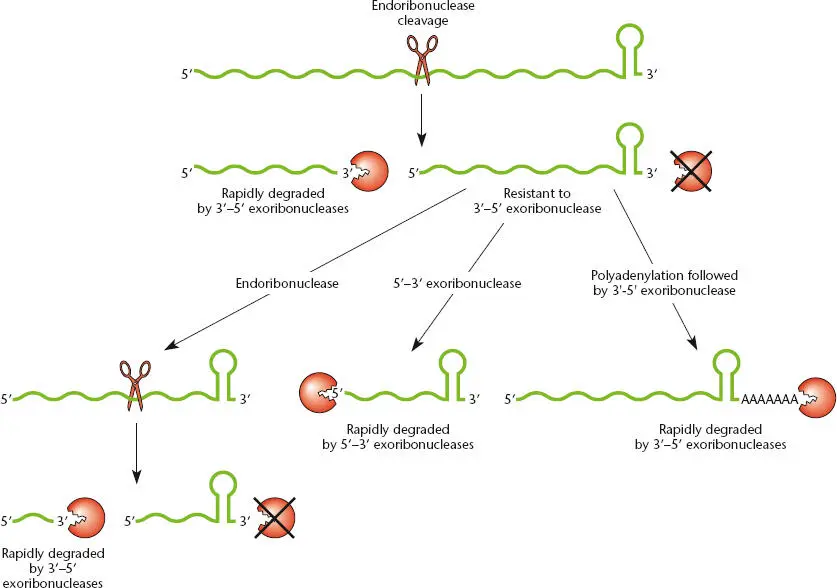
Figure 2.18 Pathways for RNA degradation. RNA transcripts that are generated by termination at a factor-independent terminator contain a hairpin at the 3′ end, which inhibits degradation by 3′-5′ exoribonucleases. Degradation is often initiated by cleavage by an endonuclease, followed by rapid exonucleolytic digestion from the new 3′ end. The stable 3′ fragment (which retains the terminator hairpin) can be cleaved again by an endoribonuclease or can be degraded by a 5′-3′ exoribonuclease in organisms that have this class of enzyme. Alternatively, poly(A) polymerase can add a poly(A) tail to the 3′ end of the RNA, which allows binding of a 3′-5′ exoribonuclease and degradation.
The Structure and Function of Proteins
Proteins do most of the work of the cell. While there are a few RNA enzymes (ribozymes), most of the enzymes that make and degrade energy sources and make cell constituents are proteins. Also, proteins contribute to much of the structure of the cell. Because of these diverse roles, there are many more types of proteins than there are types of other cell constituents. Even in a relatively simple bacterium, there are thousands of different types of proteins, and most of the DNA sequences in bacteria are dedicated to genes that encode proteins.
Unlike DNA and RNA, which consist of a chain of nucleotides held together by phosphodiester bonds between the sugars and phosphates, proteins consist of chains of 20 different amino acids held together by peptide bonds(see Figure 2.19). The peptide bond is formed by joining the amino group (NH 2) of one amino acid to the carboxyl group (COOH) of the previous amino acid. These amino acids in turn are attached to other amino acids by the same type of bond, making a chain. A short chain of amino acids is called an oligopeptide, and a long chain is called a polypeptide.
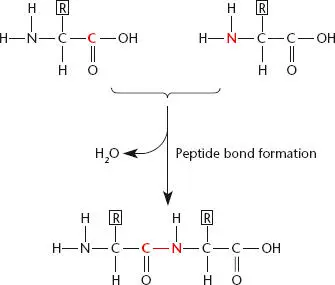
Figure 2.19 Two amino acids joined by a peptide bond. The bond connects the amino group on the second amino acid to the carboxyl group on the preceding amino acid. R is the side group of the amino acid that differs in each type of amino acid.
Like RNA and DNA, polypeptide chains have directionality and a way to distinguish the ends of the chain from each other. In polypeptides, the direction is defined by their amino and carboxyl groups. One end of the chain, the amino terminus, or N terminus, has an unattached amino group. The amino acid at this end is called the N-terminal amino acid. On the other end of the polypeptide, the final carboxyl group is called the carboxy terminus, or C terminus, and the amino acid is called the C-terminal amino acid. As we shall see, proteins are synthesized from the N terminus to the C terminus.
Protein structure terminology is the same as that for RNA structures. Proteins have primary, secondary, and tertiary structures, as well as quaternary structures. All of these are shown in Figure 2.20.
Primary structure refers to the sequence of amino acids and the length of a polypeptide. Because polypeptides are made up of 20 amino acids instead of just 4 nucleotides, as in RNA, many more primary structures are possible for polypeptides than for RNA chains. The sequence of amino acids in a polypeptide is dictated by the sequence of nucleotides in the mRNA used as the template for synthesis of that protein.
Also like RNA, polypeptides can have a secondary structure, in which parts of the chain are held together by hydrogen bonds. However, because many more types of interactions are possible between amino acids than between nucleotides, the secondary structure of a polypeptide is more difficult to predict. The two basic forms of secondary structures in polypeptides are α-helices, where a short region of the polypeptide chain forms a helix due to the interaction of each amino acid with the one before and the one after it, and β-sheets, in which stretches of amino acids interact with other stretches to form sheetlike structures ( Figure 2.20). These types of structured regions are often joined together by more flexible regions known as linkers. Computer software is available to help predict which secondary structures of a polypeptide are possible on the basis of its primary structure. However, these programs are not entirely reliable, and techniques like X-ray crystallography and nuclear magnetic resonance spectroscopy provide much more detailed information about the secondary structure of a polypeptide.
Читать дальше













