Not all methionine codons serve as initiation codons, because methionine can be found within polypeptide sequences and not only at the N-terminal end. Furthermore, the fact that some of the initiation codons (like GUG) code for amino acids other than methionine when internal to a coding region demonstrates that the presence of one codon is clearly not enough to define a TIR. These sequences also may occur out of frame or in an mRNA sequence that is not translated at all. Obviously, sequences in addition to these three bases must help to define them as a place to begin translation.
Most bacterial genes have a second component for recognizing a translational start site within the TIR. This sequence, named the Shine-Dalgarno (S-D) sequenceafter the two scientists who first noticed it, is located 5 to 10 nucleotides on the 5′ side (upstream) of the initiation codon and is complementary to a short sequence near the 3′ end of the 16S RNA. Figure 2.27shows an example of a typical bacterial TIR with a characteristic S-D sequence. By pairing with their complementary sequences on the 16S rRNA, S-D sequences help define TIRs by properly aligning the mRNA on the ribosome. However, these sequences are not always easy to identify because they can be very short and exhibit considerable variability. Moreover, not all bacterial genes have S-D sequences. The initiation codon sometimes resides at the extreme 5′ end of the mRNA (in “leaderless mRNAs”), leaving no room for an S-D sequence. In such cases, translation initiation may occur by a somewhat different mechanism (see below).
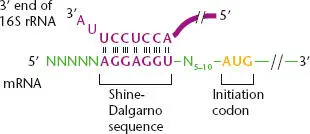
Figure 2.27 Structure of a typical bacterial translational initiation region (TIR) showing the pairing between the Shine-Dalgarno (S-D) sequence in the mRNA and a short sequence complementary to the S-D sequence that is located close to the 3′ end of the 16S rRNA. The initiation codon, typically AUG or GUG, is 5 to 10 bases downstream of the S-D sequence. N represents any base.
Because of this lack of universality, often the only way to be certain that translation is initiated at a particular initiation codon is to sequence the N terminus of the polypeptide to see if the N-terminal amino acids correspond to the codons immediately following the putative initiation codon. Protein sequencing will also reveal whether the methionine encoded by the initiation codon remains on the mature protein or is removed.
Translation initiation requires a dedicated aa-tRNA, the formylmethionine initiator tRNA (fMet-tRNA fMet). This unique aa-tRNA has a formyl group attached to the amino group of the methionine ( Figure 2.26), making it resemble a peptidyl-tRNA rather than a normal aminoacyl-tRNA because the amino group on the amino acid is blocked. This causes fMet-tRNA fMetto bind to the P site rather than the A site of the ribosome, which is an important step in initiation, as discussed below, and prevents its use as an elongator tRNA. The initiator fMet-tRNA fMetis synthesized somewhat differently from the other aminoacyl-tRNAs. This special tRNA uses the aminoacyl-tRNA synthetase of the normal tRNA Metto attach methionine to the tRNA fMet. Then, an enzyme called transformylaseadds a formyl group to the amino group of the methionine on the tRNA fMetto form fMet-tRNA fMet.
STEPS IN INITIATION OF TRANSLATION
The currently accepted view of the steps in the initiation of translation at a TIR is outlined in Figure 2.28. Initiation requires three different initiation factors, IF1, IF2, and IF3, in addition to fMet-tRNA fMet. These initiation factors interact mostly with the initiator tRNA and the P site of the 30S ribosomal subunit.
For initiation to occur, the 70S ribosome must first be separated (or dissociated) into its smaller 30S and 50S subunits. This dissociation occurs after the termination step of translation (see below). The IF3 initiation factor binds to the 30S subunit and helps to keep the subunits dissociated. Therefore, ribosomes are continuously cycling between the 70S ribosome and the 30S and 50S subunits, depending on whether they are active in translation. This is called the ribosome cycle.
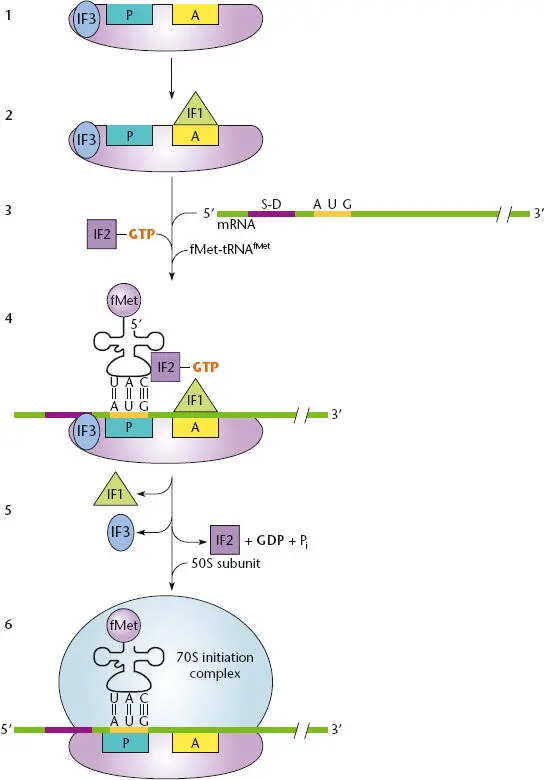
Figure 2.28 Initiation of translation. (1)The IF3 factor binds the 30S subunit to keep it dissociated from the 50S subunit during initiation. (2)IF1 binds to the A site to block the site and prevent tRNA binding. (3)A complex is formed between formyl-methionine tRNA (fMet-tRNA fMet), IF2, and guanosine triphosphate (GTP). (4)The fMet-tRNA fMet-IF2-GTP complex binds to the P site of the 30S subunit and the mRNA translational initiation region (TIR) site. (5)IF1 and IF3 are released, the cleavage of GTP on IF2 correctly positions the fMet-tRNA fMeton the P site initiation codon, and the 50S subunit binds. (6)The 70S initiation complex is ready to accept an aminoacyl-tRNA at the A site.
Once the subunits are dissociated, IF1 binds to the A site on the 30S subunit to prevent the fMet-tRNA fMetfrom inadvertently binding to this site. IF2, in conjunction with GTP, binds to fMet-tRNA fMetto form a ternary (three-member) complex, which binds to the mRNA and P site of the 30S ribosomal subunit. The initial binding of fMet-tRNA fMetdoes not depend on an initiator codon in the P site. However, IF2, with the help of IF3, adjusts the fMet-tRNA fMetand the mRNA initiator codon so that the binding becomes codon-specific. IF1 and IF3 are then released, and IF2 promotes the association of this initiation complex with the 50S subunit of the ribosome. IF2 is then released, with the cleavage of GTP to guanosine diphosphate (GDP). The newly formed 70S ribosome is now ready for translation, and another aa-tRNA can enter the A site.
Translation Initiation from Leaderless mRNAs
As mentioned above, a few mRNAs in bacteria do not have standard TIRs with leader regions containing S-D sequences. In these rare mRNAs, the initiator codon can be right at or very close to the 5′ end of the mRNA. How the ribosome recognizes such an initiator codon and initiates translation is not fully understood, but the mechanism seems to be very different from that of initiation at a normal TIR. There is some evidence that a complex first forms between fMet-tRNA fMet, IF2, and the small subunit of the ribosome. This complex may then help recognize the initiation codon in the absence of upstream sequences to help distinguish the initiation codon. Other evidence suggests that the 70S ribosome itself recognizes the leaderless initiator codon.
TRANSLATION INITIATION IN ARCHAEA AND EUKARYOTES
Translation initiation in the archaea is similar to that in the bacteria. Like bacteria, archaea use well-defined TIRs with leader sequences and formylmethionine for initiation of translation. In contrast, eukaryotes do not seem to use special sequence elements and instead usually use the first AUG from the 5′ end of the mRNA as the initiation codon. Sequences around this initiator AUG may also be important for its recognition, and secondary structure in the mRNA may mask other AUG sequences that could potentially be used as initiator codons. Although eukaryotes have a special methionyl tRNA that responds to the first AUG codon, called Met-tRNA i, the methionine attached to the eukaryotic initiator tRNA is never formylated. As in bacteria, however, the first methionine is usually removed by an aminopeptidase after the protein is synthesized. Eukaryotes and archaea also seem to use many more initiation factors and elongation factors than do bacteria. The archaea use formylated methionine and S-D sequences like bacteria, but their initiation factors are more similar to those in eukaryotes. Although the exact roles of most of these initiation factors are unknown in archaea, many are obviously related to the initiation and translation factors of bacteria. It therefore appears that the mechanism of translation initiation in the archaea is a sort of hybrid between that in bacteria and that in eukaryotes.
Читать дальше













