bShorter product of the dnaX gene produced by translational frameshifting (see chapter 2).
Enzymes that degrade DNA strands by breaking the phosphodiester bonds are just as important in replication as the enzymes that polymerize DNA by forming phosphodiester bonds between the nucleotides. These bondbreaking enzymes, called nucleases, can be grouped into two major categories. One type can initiate breaks in the middle of a DNA strand and so are called endonucleases, from a Greek word meaning “within,” and the other type can remove nucleotides only from the ends of DNA strands and so are called exonucleases, from a Greek word meaning “outside.” A special type of endonuclease activity, called a flap endonucleaseactivity, is involved in primer removal by DNA polymerase I. The flap endonuclease activity appears to be common to all organisms for removing RNA primers. In E. coli , DNA polymerase I displaces the RNA primer, making a flap-like structure, and then the flap endonuclease activity of Pol I cleaves away the oligonucleotide as indicated ( Figure 1.9). The exonucleasescan be subdivided into two groups. Some exonucleases can degrade only from the 3′ end of a DNA strand, degrading DNA in the 3′-to-5′ direction. These are called 3′ exonucleases; one example of their activity is their role in the editing function associated with DNA polymerases I and III, which is discussed below. Other exonucleases, called 5′ exonucleases, degrade DNA strands only from the 5′ end.
DNA ligasesare enzymes that form phosphodiester bonds between the ends of separate presynthesized chains of DNA. This important function cannot be performed by any of the known DNA polymerases. During replication, ligase joins the 5′ phosphate at the end of one DNA chain to the 3′ hydroxyl at the end of another chain to make a longer, continuous chain ( Figure 1.8).
Replication of large DNAs requires many functions that reside in proteins separate from the subunit used for polymerizing the chain of nucleotides. These functions include the coordination of multiple DNA polymerases and tethering of these components to the template DNA strands as a moving production platform. DNA polymerase III is the major DNA replication protein in E. coli responsible for polymerizing the new complementary DNA strands, and it functions with multiple DNA polymerase accessory proteinsthat travel along the template strand with the molecule of DNA polymerase III. The term DNA polymerase III holoenzymecan be used to describe the entire complex of proteins. The various subunits and subassemblies of the DNA polymerase III holoenzyme were originally identified from fractionation procedures and were designated by Greek letters (Table 1.1).
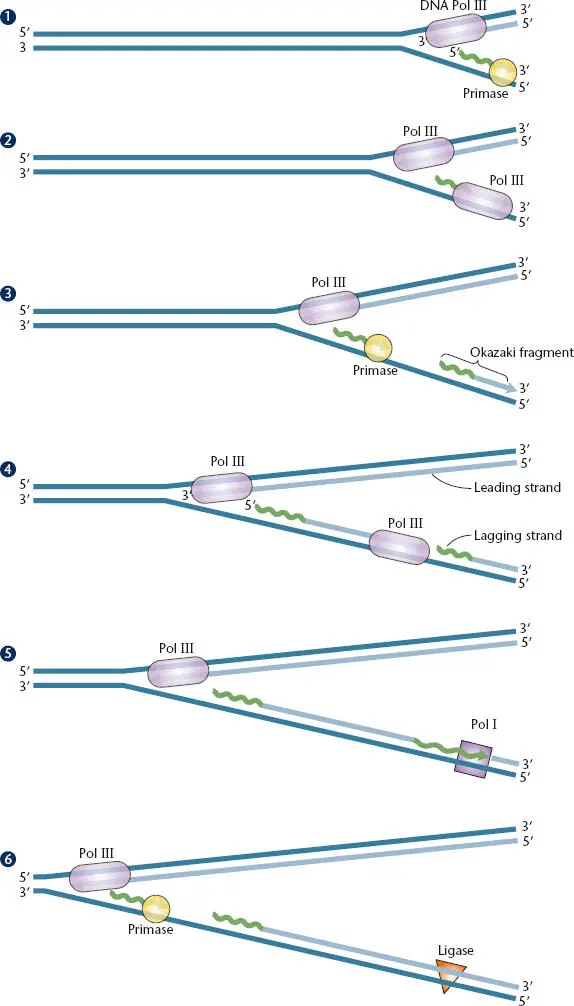
Figure 1.8 Discontinuous synthesis of one of the two strands of DNA during chromosome replication. (1)DNA polymerase (Pol) III replicates one strand, and the primase synthesizes RNA on the other strand in the opposite direction. (2)Pol III extends the RNA primer to synthesize an Okazaki fragment. (3)The primase synthesizes another RNA primer. (4)Pol III extends this primer until it reaches the previous primer. (5)Pol I removes the first RNA primer and replaces it with DNA. (6)DNA ligase seals the nick to make a continuous DNA strand, and the process continues. The strand that is synthesized continuously is the leading strand; the strand that is synthesized discontinuously is the lagging strand.
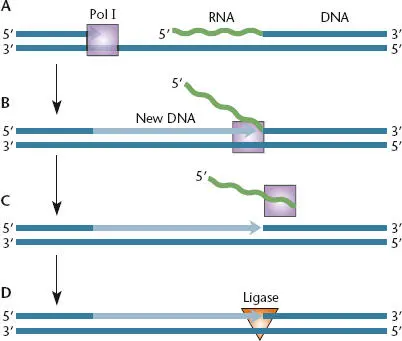
Figure 1.9 DNA polymerase I can remove an RNA primer by using strand displacement and endonuclease activity. (A)The DNA strand produced by DNA polymerase III holoenzyme is extended by DNA polymerase I until it encounters a previously synthesized RNA primer. (B)During the process of DNA replication, DNA polymerase I displaces the RNA primer. (C)An endonuclease activity in DNA polymerase I is used to cleave off the RNA primer. (D)Ligase joins the new Okazaki fragment to the previous Okazaki fragment to allow a contiguous DNA.
One of the DNA polymerase accessory proteins forms a ring around the template DNA strand and is responsible for keeping DNA polymerase from falling off. Because this ring slides freely over double-stranded DNA and will not easily come off the DNA, it is also referred to as a sliding clamp, or β clamp. The β clamp provides the foundation of the mobile platform for DNA replication, allowing it to continue for long distances without being released. In bacteria, the β clamp is a product of the dnaN gene, where two head-to-toe molecules form the ring around the DNA. While it was first isolated as part of the DNA polymerase III holoenzyme, the β clamp protein is important for multiple DNA transactions.
A special subcomplex within DNA polymerase III is the clamp loader, which is responsible for loading β clamp proteins onto the DNA. The clamp-loading complex is also responsible for tethering proteins across the DNA replication fork; the clamp loader binds the DNA polymerases on both DNA template strands and the enzyme responsible for separating the DNA strands (see below). The clamp loader is a complicated structure that consists of one γ and two τ proteins and one each of δ and δ', which form a five-sided structure, and two additional proteins, χ and ψ. The clamp loader complex is also responsible for removing β clamps. The rate of clamp removal allows many β clamps to reside on the DNA for a period of time after the replication fork passes (see Moolman et al., Suggested Reading). β clamps temporarily left behind on the newly replicated DNA play a role in helping to recruit other proteins responsible for various replication and repair functions described in this and other chapters.
Replication of Double-Stranded DNA
Additional complications of DNA replication come from the fact that the DNA is double stranded and the strands are antiparallel. The replication of all bacterial chromosomes begins at one point, called the origin of replication, with the replication enzymes moving in opposite directions from this point along the chromosome. In this process, both strands of DNA are replicated at the same time with a coordinated set of proteins. Replicating the antiparallel strands is further complicated by the abovementioned fact that DNA polymerases can replicate only in the 5′-to-3′ direction. Therefore, one DNA strand is replicated in the same direction that the replication fork is moving, and in theory, replication of this strand could continue without the need for reinitiating in a process called leading-strandDNA synthesis. However, replication of the other DNA strand occurs in the opposite direction from the progression of the replication machinery. Replication of this strand must continually be reinitiated in a process known as lagging-strandDNA synthesis. Replication of double-stranded DNA requires coordination between multiple holoenzyme subunits and DNA polymerases, as well as a host of other replication proteins.
Читать дальше













