The deoxynucleoside triphosphate dTTP is synthesized by a somewhat different pathway from the other three. The first step is the same. Ribonucleotide reductase synthesizes the nucleotide dUDP (deoxyuridine diphosphate) from the ribose UDP. However, from then on, the pathway differs. A phosphate is added to make dUTP, and the dUTP is converted to dUMP by a phosphatase that removes two of the phosphates. This molecule is then converted to dTMP by the enzyme thymidylate synthetase, using tetrahydrofolate to donate a methyl group. Kinases then add two phosphates to the dTMP to make the precursor dTTP.
Replication of the Bacterial Chromosome
Once the precursors of DNA replication are synthesized, they must be polymerized into long double-stranded DNA molecules. A very large complex of many enzymes assembles on the DNA and moves along the DNA, separating the strands and making a complementary copy of each of the strands. Thus, two strands of DNA enter the complex and four strands emerge on the other side, forming a branched structure. Each of the emerging branches contains one old “conserved” strand and one new strand, hence the name semiconservation replication. This branched structure where replication is occurring is called the replication fork. In this section, we discuss what is happening in the replication fork, including the mechanisms used to overcome obstacles to the progression of the replication fork on the chromosome.
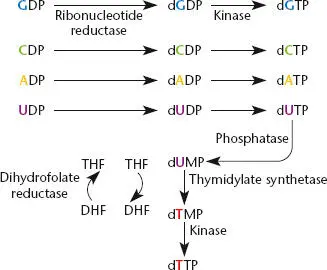
Figure 1.5 The pathways for synthesis of deoxynucleotides from ribonucleotides. Some of the enzymes referred to in the text are identified. THF, tetrahydrofolate; DHF, dihydrofolate.
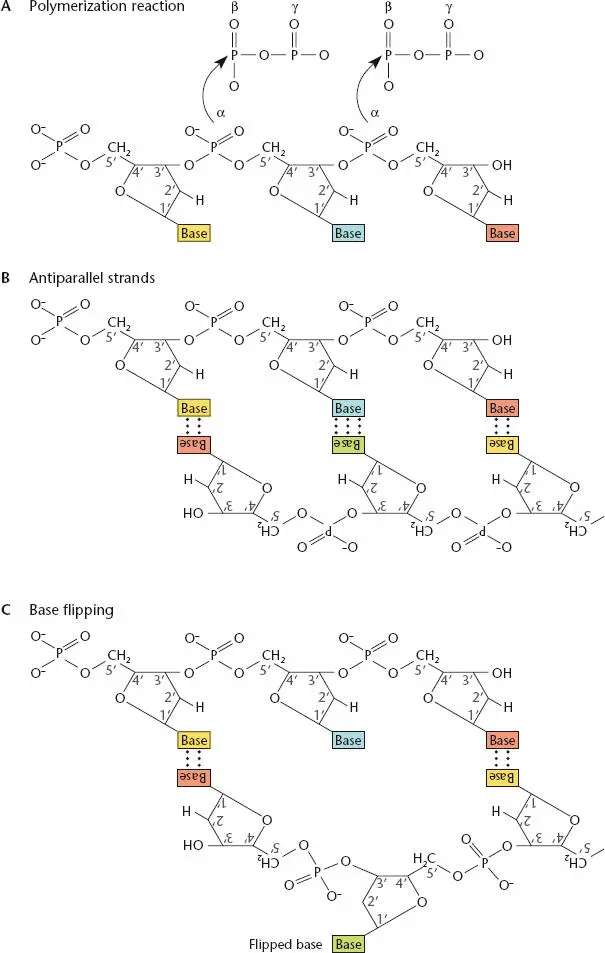
Figure 1.6 Features of DNA. (A)Polymerization of the deoxynucleotides during DNA synthesis. The β and γ phosphates of each deoxynucleoside triphosphate are cleaved off to provide energy for the polymerization reaction. (B)The strands of DNA are antiparallel. (C)A single base can be flipped out from the double helix, which could be important in recombination and repair.
The properties of the DNA polymerases, the enzymes that join the deoxynucleotides together to make the long chains, are the best guides to an understanding of the replication of DNA. These enzymes make DNA by linking one deoxynucleotide to another to generate a long chain of DNA. This process is called DNA polymerization, hence the name DNA polymerases.
Figure 1.6shows the basic process of DNA polymerization by DNA polymerase. The DNA polymerase attaches the first phosphate (the α phosphate) of one deoxynucleoside triphosphate to the 3′ carbon of the sugar of the next deoxynucleoside triphosphate, in the process releasing the last two phosphates (the β and γ phosphates) of the first deoxynucleoside triphosphate to produce energy for the reaction. Then the α phosphate of another deoxynucleoside triphosphate is attached to the 3′ carbon of this deoxynucleotide, and the process continues until a long chain is synthesized.
DNA polymerases also need a template strandto direct the synthesis of the new strand ( Figure 1.7). As mentioned in “Base Pairing” above, complementary base pairing dictates that wherever there is a T in the template strand, an A is inserted in the strand being synthesized and so forth according to the base-pairing rules. The DNA polymerase can move only in the 3′-to-5′ direction on the template strand, linking deoxynucleotides in the new strand in the 5′-to-3′ direction. When replication is completed, the product is a new double-stranded DNA with antiparallel strands, one of which is the old template strand and one of which is the newly synthesized strand.
There are two DNA polymerases that participate in normal DNA replication in Escherichia coli ; they are called DNA polymerase III and DNA polymerase I (Table 1.1). DNA polymerase III is a large protein complex in which the enzyme that polymerizes nucleotides works with numerous accessory proteins. In E. coli and other bacteria, DNA polymerase III is responsible for the bulk of DNA replication on both DNA strands. As discussed below, DNA polymerase I has a number of features that are important because replication is continually reinitiated on one of the DNA strands. It also plays a role in DNA repair, as discussed in chapter 10. Table 1.1 lists many of the DNA replication proteins, the genes encoding them, and their functions.
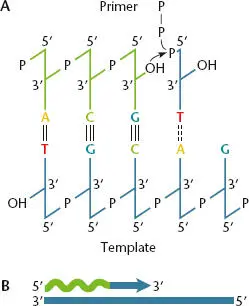
Figure 1.7 Functions of the primer and template in DNA replication. (A)The DNA polymerase adds deoxynucleotides to the 3′ end of the primer by using the template strand to direct the selection of each base. (B)Simple illustration of 5′-to-3′ DNA synthesis. The wavy green line represents the primer.
One type of enzyme, called a primase, is required during DNA replication because DNA polymerases cannot start the synthesis of a new strand of DNA; they can only attach deoxynucleotides to a preexisting 3′ OH group. The 3′ OH group to which DNA polymerase adds a deoxynucleotide is called the primer( Figure 1.7). The requirement for a primer for DNA polymerase creates an apparent dilemma in DNA replication. When a new strand of DNA is synthesized, there is no DNA upstream (i.e., on the 5′ side) to act as a primer. Primase makes small stretches of RNA that are complementary to the template strand, which are in turn used to initiate, or prime, polymerization of a new strand of DNA ( Figure 1.8). In some special situations, an RNA primer for DNA replication can be made by the RNA polymerase used for information processing (i.e., the RNA polymerase that makes all the other RNAs, including mRNA, tRNA, and rRNA; see chapter 2). Unlike DNA polymerase, primase and RNA polymerases do not require a primer to initiate the synthesis of new strands. During DNA replication, a special enzyme activity recognizes and removes the RNA primer (see below).
Table 1.1Proteins involved in Escherichia coli DNA replication
| Protein |
Gene |
Function |
| DnaA |
dnaA |
Initiator protein; primosome (priming complex) formation |
| DnaB |
dnaB |
DNA helicase |
| DnaC |
dnaC |
Delivers DnaB to replication complex |
| SSB |
ssb |
Binding to single-stranded DNA |
| Primase |
dnaG |
RNA primer synthesis |
| DNA ligase |
lig |
Sealing DNA nicks |
| DNA gyrase |
|
Supercoiling |
| α |
gyrA |
Nick closing |
| β |
gyrB |
ATPase |
| DNA Pol I |
polA |
Primer removal; gap filling |
| DNA Pol III (holoenzyme) |
|
|
| α |
dnaE |
Polymerization |
| ε |
dnaQ |
3′-to-5′ editing |
| RNase H |
rnhA |
Can aid in RNA primer removal |
| θ |
holE |
Present in core (αεθ) |
| β |
dnaN |
Sliding clamp |
| τ a |
dnaX |
Organizes complex; joins leading and lagging DNA Pol III |
| γb |
dnaX |
Binds clamp loaders and single-strand-binding protein |
| δ |
holA |
Clamp loading |
| δ' |
holB |
Clamp loading |
| χ |
holC |
Binds single-strand-binding protein |
| φ |
holD |
Holds χ to the clamp loader |
aFull-length product of the dnaX gene.
Читать дальше














