The genetic architecture of quantitative traits entails the number of QTLs that influence a quantitative trait, the number of alleles that each QTL possesses, the frequencies of the alleles in the population, and the influence of each QTL and its alleles on the quantitative trait. Identifying and characterizing QTLs will provide a basis for selecting and improving plant species. The summation of QTL studies indicates that QTL alleles with large effects are rare; most quantitative traits are controlled by many loci with small effects.
Researchers commonly use one of two fundamental approaches to design and study quantitative traits. In what is called the top‐down approach, they start with the trait of interest and then attempt to draw inferences about the underlying genetics from examining the degree of trait resemblance among related subjects. It is usually the first step taken to determine if there is any evidence for a genetic component. It is also described as the unmeasured genotype approach because it focuses on the inheritance pattern without measuring any genetic variations. Typical statistical analyses employed in this approach are heritabilityand segregational analysis.
In the second approach, the bottom‐up (measured) approach, researchers actually measure QTLs and use the information to draw inferences about which genes might have a role in the genetic architecture of a quantitative trait. Typical statistical analyses employed in this approach are linkage analysisand association analysis.The second approach is becoming more assessable with the advent of newer, more efficient, and less expensive technologies to measure QTLs. These technologies include DNA microarrays and protein mass spectrometry. They allow researchers to quantitatively measure the expression levels of thousands of gene simultaneously, and thereby study gene expression at both the ribonucleic acid (RNA) and protein levels as a quantitative trait.
4.3.1 Effects of QTL on phenotype
QTLs influence quantitative trait phenotype in various ways. They can influence quantitative trait levels (quantitative trait means can be different among different genotypes). Most of the statistical methods used for studying quantitative traits are based on genotypic means. The variation in phenotypic values may also vary among genotypes. Further, QTLs also may affect the correlation among quantitative traits as well as the dynamics of traits (the change in phenotype over a period of time may be due to variations in a QTL).
QTL alleles have context‐dependent effects. Their effects may differ in magnitude or direction in different genetic backgrounds, different environments, or between male and females (i.e. genotype × genotype interaction – epistasis; genotype × environment interactions; genotype × sex interaction). These context‐dependent effects are very common and play a significant role in genetic architecture, but they are very difficult to detect and characterize. In addition to meaning the masking of genotypic effects at one locus by genotypes of another locus, epistasis in quantitative genetics also refers to any statistical interaction between the genotypes at two or more loci. It is common between mutations that affect the same quantitative trait. Epistatic effects can be as large as main QTL effects; they can also occur in opposite directions between different pairs of interacting loci and between loci with having significant main effects on the trait of interest. Epistasis has been found between closely linked QTLs and also polymorphisms at a single locus.
Pleiotropy, the effect of a gene on more than one phenotype, is important in the genetics of QTLs. In a narrow sense, pleiotropy can mean the effect of a particular allele on more than one phenotype, and is the reason for the stable genetic correlations between quantitative traits, if the effects at multiple loci affecting the same trait are in the same direction. Understanding of pleiotropic connects between quantitative traits helps in predicting the correlated responses to artificial selection and assessing the contribution of new mutations to standing variation for quantitative traits. Pleiotropy is known to occur even between traits that are not functionally related. Consequently, the pleiotropic effects of different genes that affect pairs of traits are usually not in the same direction and do not result in significant genetic correlations between traits.
4.3.2 Molecular basis of quantitative variation
The causal molecular variants that affect QTL are called quantitative trait nucleotide s (QTN s).The distribution of QTN allele frequencies can indicate the nature of the selective forces operating on the trait. Inference of QTN allele frequencies is limited to association mapping (see Chapter 20) designs in which all the variants in a candidate gene or gene region have been identified. QTNs allow researchers to map phenotype to genotype in the absence of biological context. QTNs consist of two components: eQTL ( expression quantitative trait loci )and QTT ( quantitative trait transcripts )( Figure 4.8) .
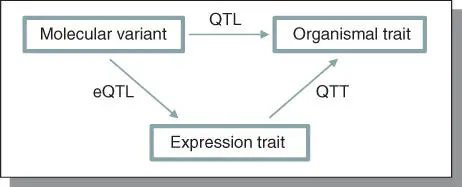
Figure 4.8Systems genetics of complex traits: An integrative framework showing the relationship between DNA sequence variation and quantitative variation for gene expression and an organismal phenotype. QTNs allow researchers to map phenotype to genotype in the absence of biological context. In order to gain this context, they need to describe the flow of information from DNA to the organismal phenotype through RNA intermediates, proteins and other molecular endophenotypes.
The eQTL is a region of the genome containing one or more genes that affect variation in gene expression, which is identified by linkage to polymorphic marker loci. It is technically a marriage of high‐throughput expression profiling technology and QTL analysis. QTT is a transcript for which variation in its expression is correlated with variation in an organismal level quantitative trait phenotype. Numerous (even several hundreds) of QTT are believed to be associated with any single quantitative trait phenotype. Further, these QTT are genetically correlated.
Systems genetics(also called genetical genomics) is considered to be the third in the developmental stages of quantitative genetics, after classical and molecular quantitative genetics in that order of development. This new paradigm is instigated by the fact that associating DNA sequence variation with variation of the phenotype of an organism skips all of the intermediate steps (e.g. the intermediate molecular phenotypes like transcript abundance which are quantitative traits and vary genetically in populations) in the chain of the causation from genetic perturbation to phenotypic variation. Systems genetics detects variation in phenotypic traits and integrates them with underlying genetic variation. According to Mackay and colleagues, this approach to QTL analysis integrates DNA sequence variation, variation in transcript abundance and other molecular phenotypes, and variation in the phenotypes of the organism in linkage or association mapping population, thereby allowing the researcher to interpret quantitative genetic variation in terms of biologically meaningful causal networks of correlated transcripts. In other words, it can be used to define biological networks and to predict molecular interactions by analyzing transcripts with expressions that co‐vary within genetic populations. The approach can be used to analyze effects of genome‐wide genetic variants on transcriptome‐wide variation in gene expression.
Читать дальше













