Self‐assembly of DNA is one of the most popular techniques used to create a variety of two‐dimensional and three‐dimensional structures. The DNA origami is an excellent example of self‐assembly of DNA, which was introduced by Rothemund [14]. The procedure of DNA origami is shown in Figure 2.8. DNA origami is generated from a thousand base pair long ssDNA, known as a scaffold. Some shorter and linear ssDNA called staples are mixed with the scaffold to create DNA origami. For this purpose, the solution mixture is first heated to 95 °C. The solution is then cooled down to room temperature from 95 °C; throughout the cooling, the staples bind to the scaffold through Watson–Crick complementary base pairing. The scaffold then becomes a static structure or pattern of DNA. The sequence of DNA staples results in the formation of various shapes, for example, squares, smiley faces, cube, hexagon, star, and many more.
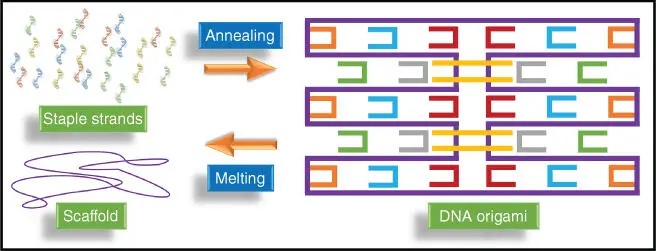
Figure 2.8DNA origami is a group of linked dsDNA. The structure consists of one scaffold and many staple strands that are complementary to one or two domains on the scaffold.
Based on the structure or pattern of the DNA origami, it has multiple applications in different fields of biology, chemistry, physics, computer science, and materials science. Broad application of DNA origami can be seen as in molecular robotics [15,16], DNA walkers [17], protein function [18,19] and structure determination [20,21], single‐molecule force spectroscopy [22,23], nanopore construction [24–26], drug delivery [27,28], nucleic acid analysis [29–31], enzymatic nanostructure formations [32,33], and many more.
2.2.8 DNA‐Based Data Storage
DNA carries all the genetic information such as the color of eyes and hairs in the form of A, T, G, and C, which is transferred from one generation to another. Inspired by this, the researchers are now looking at the capabilities of the DNA to store the data. Today we are dealing with a large amount of data that is increasing day by day. DNA appears to be the right choice as an alternative for storing the information. In DNA‐based data storage, first, the digital information present in the form of bits is encoded in DNA. This encoded information is then processed and decoded back to original binary data.
Church et al. [34] performed an experiment for storing 5.27 MB data generated from his book (contains 53 426 words, 11 images, and 1 JavaScript program) on the DNA. A simple encoding of one bit to one base is used to represent the data on the DNA. The result of this experiment clearly showed that DNA is a good material for storing digital information in addition to other storage media. Goldman et al. [35] stored over five million bits of digital information of the DNA that is later retrieved and reproduced the information with an accuracy between 99.99% and 100%.
Researchers are now looking for high data storage with high accuracy in recovery. Blawat et al. [36] stored and recovered 22 MB of a MPEG compressed movie from DNA with zero errors. Erlich and Zielinski [37] reported another encoding method that can store the 215 petabytes digital data in one gram of DNA. Recently, Organick et al. [38] demonstrated approximately 220 MB digital data storage with random access on the DNA with successful retrieval from it. In 2019, researchers from Microsoft and the University of Washington have demonstrated a fully automated system to encode and decode data in DNA [39].
Though the use of DNA for computing can have the benefit of performing millions of operations simultaneously with very high energy efficiency, it also has several challenges. The amount of DNA required is substantial even for a simple formulation. Therefore, solving the large size problems becomes impractical owing to the requirement of a large amount of DNA. Unlike the traditional silicon‐based computers in which memory reallocation is performed readily, reuse of DNA material is challenging in DNA computing as specific designs of DNA are required.
The success of DNA computing procedures is based on error‐free operations of biochemical steps involved. However, the practical operations do involve experimental errors that increase with the increase in the number of steps. Further, the operations involve human interventions during the process. Therefore, solving the bigger size formulations also involves the higher probability of missing the correct answers. Further, increase in formulation size requires extracting the correct solution from the pool involving large number of incorrect solutions. Therefore, the extraction efficiency decreases with increase in formulation size. Also, the large amount of DNA representing the incorrect solutions is discarded as waste. Complete automation of all the biochemical steps is required for building a reliable DNA computer.
Another challenge for DNA computing is its application to real‐world problems. Since the data of every problem has to be represented in the form of DNA, the design of DNA is specific to each problem and cannot be used for other problems. Further, for error‐free biochemical operations, the DNA designs should have specific GC content with unique (noncomplementary) nucleotide sequence and should lead to a specific structure (i.e. hairpin or linear formation). These requirements reduce the designing flexibility and therefore restrict the application to bigger size formulations. Moreover, real‐world problems often involve continuous search spaces with multiple optimal solutions. For such problems, the existing DNA computing procedures that are originally developed for solving the combinatorial problems involving the discrete search space need to be modified.
In conclusion, DNA computing shows great potential and has many advantages in the field of computing and data storage over conventional computing, primarily due to its ability to perform millions of calculations simultaneously using molecules. Despite this, the DNA computer is far from matching the reliability of conventional silicon‐based computer owing to several challenges such as poor scaling and limited ability to handle real‐world problems. The comparative analysis of existing DNA computing and data storage models illustrated their pros and cons, which is opening up new directions in materials science and biomedical applications.
This chapter is a part of the PhD thesis titled “Computing using Biomolecules” of Mr. Deepak Sharma, which is under consideration for the award of PhD degree at Indian Institute of Technology Delhi, India.
1 1 Moore, G.E. (1975) Progress in digital integrated circuits. Electron Devices Meeting, 11–13.
2 2 Adleman, L.M. (1994). Molecular computation of solutions to combinatorial problems. Science266 (5187): 1021–1024.
3 3 Lipton, R.J. (1995). DNA solution for hard computational problems. Science268 (5210): 542–545.
4 4 Smith, L.M., Corn, R.M., Condon, A.E. et al. (1998). A surface‐based approach to DNA computation. J Comput Biol5 (2): 255–267.
5 5 Sakamoto, K., Gouzu, H., Komiya, K. et al. (2000). Molecular computation by DNA hairpin formation. Science288 (5469): 1223–1226.
6 6 Liu, Q., Wang, L., Frutos, A.G. et al. (2000). DNA computing on surfaces. Nature403 (6766): 175–179.
7 7 Braich, R.S., Chelyapov, N., Johnson, C. et al. (2002). Solution of a 20‐variable 3‐SAT problem on a DNA computer. Science296 (5567): 499–502.
8 8 Kahan, M., Gil, B., Adar, R., and Shapiro, E. (2008). Towards molecular computers that operate in a biological environment. Physica D237 (9): 1165–1172.
Читать дальше













