Cheanyeh Cheng - Enzyme-Based Organic Synthesis
Здесь есть возможность читать онлайн «Cheanyeh Cheng - Enzyme-Based Organic Synthesis» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Enzyme-Based Organic Synthesis
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:3 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 60
- 1
- 2
- 3
- 4
- 5
Enzyme-Based Organic Synthesis: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Enzyme-Based Organic Synthesis»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
An insightful exploration of an increasingly popular technique in organic chemistry Enzyme-Based Organic Synthesis
Enzyme-Based Organic Synthesis
Enzyme-Based Organic Synthesis
Enzyme-Based Organic Synthesis — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Enzyme-Based Organic Synthesis», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
Since proteins are large macromolecules, the complexity of their 3D structure cannot be described easily like small molecules. Therefore, four levels of structure are used to define the complete 3D structure of protein. The primary structure is the amino acid sequence of a polypeptide chain that describes the order of all covalent bonds, mainly peptide bonds and disulfide bonds, linking amino acid residue in the polypeptide as illustrated in Scheme 1.1. The importance of primary structure is in determining the secondary, tertiary, quaternary structures of proteins, and thus their biological functions, which can be demonstrated by the hereditary disease sickle‐cell anemia of human.
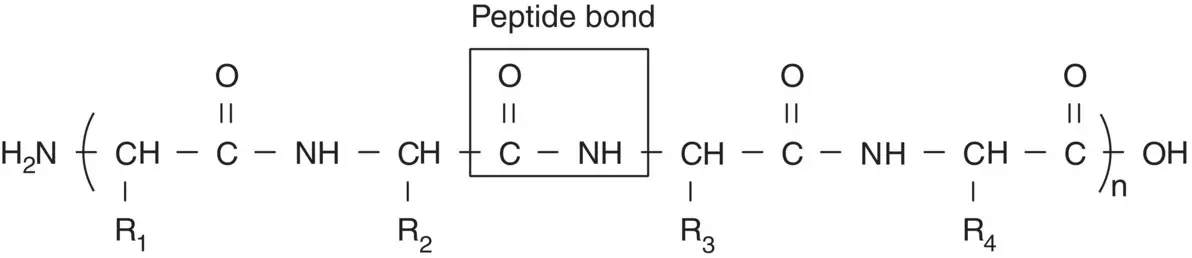
Scheme 1.1 The primary structure of a polypeptide chain linked by the peptide bond shows the sequence of amino acids.
Part of the very long chain polypeptide can be coiled or folded into units by amino acid residues within a short distance to form recurring structural patterns of secondary structure such as the α‐helix of α‐keratin. The helix is a part of the tertiary structure that is the overall 3D arrangement or folding of a polypeptide. An example of the tertiary structure is myoglobin, a globular protein with 153 amino acid residues. The secondary structure refers to the spatial arrangement of amino acid residues that are adjacent in the primary structure, whereas tertiary structure includes longer‐range aspects of the primary structure. When a protein has two or more polypeptide subunits that are associated with each other or one another, their arrangement in space is referred to as quaternary structure. Hemoglobin consisting of four polypeptide subunits is the most well‐known protein with a complex quaternary structure.
1.2.2 Catalytic Function
The folding of long polypeptide chain to form tertiary or quaternary structure is caused by chemical or physical forces such as disulfide linkage, hydrogen bonding, acid–base interaction (salt bridge), and hydrophobic interaction. Folding of polypeptides into two or more stable globular units is called domains. Different domains often play distinct functions, such as binding molecules or interaction with other proteins. A molecule bound reversibly by a protein is called ligand. The site on the protein that binds the ligand is called the binding site. When a protein binds a ligand, the 3D structure of protein is often caused by a conformational change to permit a tighter binding to the ligand. This kind of binding with structural adaption between protein and ligand is called induced fit mechanism. Enzymes have catalytic function that binds and chemically rapidly transforms other molecules. For enzyme‐catalyzed reaction, the molecule bound and acted by the enzyme is called substrate; the binding site is called the active site or catalytic site.
Enzyme is an efficient catalyst and is responsible for thousands integrated chemical reactions of the biological process occurred in the living system. Just like the usual inorganic catalyst, enzymes catalyze a reaction by lowering the transition state energy (the activation energy) of the activated complex and by raising the ground state energy. On the other hand, the catalysis of enzymes, not like simple inorganic catalysts, proceeds by forming several transition states and each with low activation energy instead of one activated complex of greater activation energy. The rate of enzyme‐catalyzed reaction for a simple one enzyme, one substrate and one product system with the following mechanism was studied by Michaelis and Menten in 1913 ( Scheme 1.2). In this mechanism, enzyme (E) binds the substrate (S) to form an enzyme–substrate (ES) complex and subsequently the ES breaks down to the product (P).
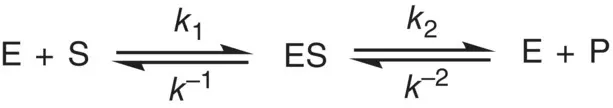
Scheme 1.2 The proposed enzyme reaction mechanism by Michaelis and Menten.
According to this mechanism, the enzyme‐catalyzed reaction rate equation called Michaelis–Menten equation ( Eq. 1.1) was derived by Michaelis and Menten with the second step as the rate‐limiting step and derived by Briggs and Haldane using steady‐state assumption. The term V 0is the initial rate, V maxis the maximum reaction rate, [ S ] is the substrate concentration, and K mis the Michaelis constant.
(1.1) 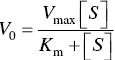
For a more common case, enzyme product complexes (EP) release, EP → E + P, is the rate‐limiting step, which is described as the reaction Scheme 1.3.
Therefore, a more general rate constant called the turnover number, k cat, is defined to describe the limiting rate of any enzyme‐catalyzed reaction at saturation, that is, V max= k cat[ E t]. In this situation, the Michaelis–Menten equation becomes Eq. 1.2
(1.2) 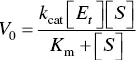
The turnover numbers of several enzymes are given in Table 1.1.
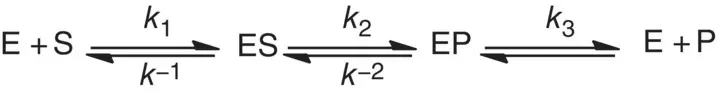
Scheme 1.3 A generalized enzyme‐catalyzed reaction.
Table 1.1 Turnover numbers for some enzymes.
| Enzyme | Turnover number k cat(s −1) |
|---|---|
| Catalase | 40 000 000 |
| Carbonic anhydrase | 400 000 |
| Acetylcholinesterase | 140 000 |
| β‐Lactamase | 2000 |
| Fumarase | 800 |
| β‐Galactosidase | 208 |
| Phosphoglucomutase | 21 |
| Tryptophan synthetase | 2 |
| RecA protein (an ATPase) | 0.4 |
1.3 Cofactors and Coenzymes
Enzymes have protein nature and molecular weights ranging from about 12 000 to over 1 million. The large molecule of enzymes is flexible for binding natural and unnatural substrates at their active site. The active site contains moieties consisted with amino acid residues. Although the activity of some enzymes requires no chemical groups other than their amino acid residues, others require an additional chemical component called cofactor. A cofactor, also called a coenzyme, is either one or more inorganic ions, such as Fe 2+, Mg 2+, Mn 2+, or Zn 2+( Table 1.2) [9], or an organic or metallo‐organic molecule. Coenzyme are often derived from vitamins and organic nutrients required in small amounts in the diet ( Table 1.3) [9]. The cofactor binds to the active site, in some cases covalently and in others noncovalently, which serves as transient carriers of redox equivalents, such as NAD(P)H or chemical energy (ATP) and is essential for the catalytic action of those enzymes that require cofactors.
Table 1.2 Some inorganic metal ions as cofactor of enzymes.
Source: Based on Nelson and Cox [9].
| Fe 2+or Fe 3+ | Cytochrome oxidase, catalase, peroxidase |
| K + | Pyruvate kinase |
| Mg 2+ | Hexokinase, pyruvate kinase, enolase |
| Mn 2+ | Arginase, ribonucleotide reductase |
| Ni 2+ | Urease |
| Zn 2+ | Carbonic anhydrase, alcohol dehydrogenase, carboxypeptidases A and B |
Table 1.3 Some coenzymes as transient carriers of specific atoms or functional groups.
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Enzyme-Based Organic Synthesis»
Представляем Вашему вниманию похожие книги на «Enzyme-Based Organic Synthesis» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Enzyme-Based Organic Synthesis» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.












