1 ...6 7 8 10 11 12 ...29 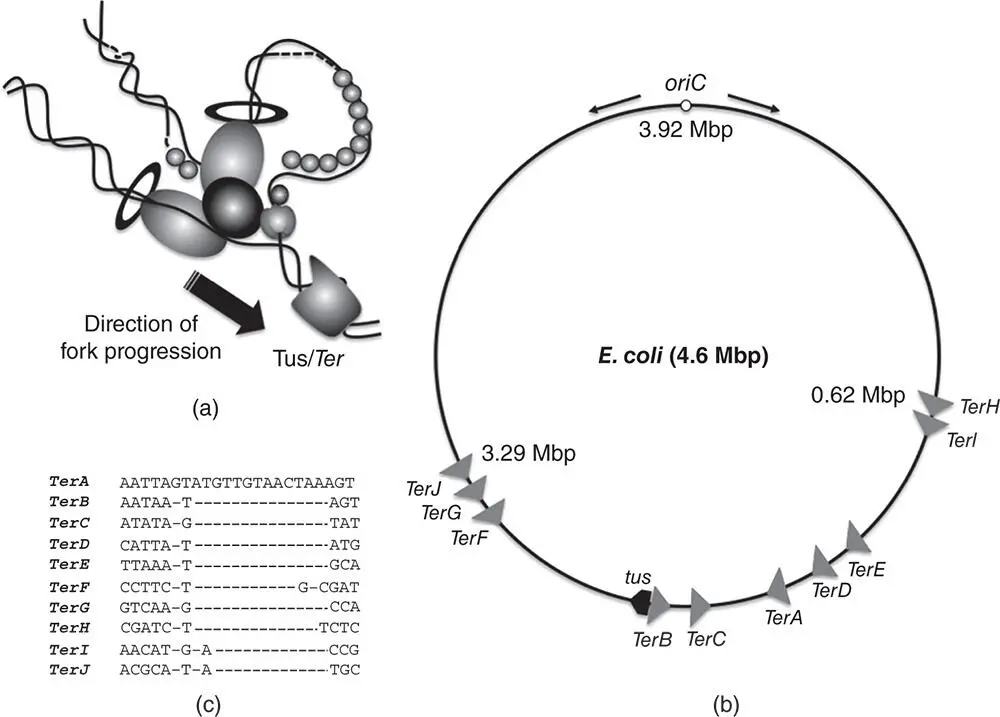
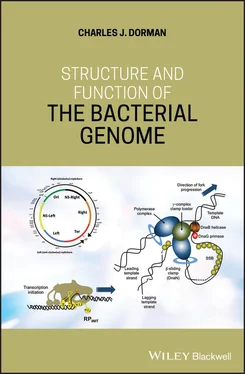
Figure 1.7Termination of chromosome replication in E. coli . (a) The moving replisome encounters an appropriately oriented Tus/ Ter nucleoprotein complex and the interaction between Tus and the DnaB helicase halts replisome movement, leading to the termination of chromosome replication. (b) The 4.6 Mb chromosome of E. coli is shown, indicating the relative positions and orientations of the Ter sequences (grey arrowheads) with respect to one another and oriC and the tus gene. The black arrows on either side of oriC indicate the bidirectional nature of E. coli chromosome replication. Ter sites aligned with the direction of replication are in the permissive orientation and will allow the replication fork to pass; those oriented against the direction of fork movement are in the non‐permissive configuration and will halt fork movement if bound by Tus. The promoter of the tus gene overlaps the TerB sequence, resulting in negative autoregulation of tus transcription by the Tus protein. (c) An alignment of the DNA sequences of the Ter elements, showing the high degree of sequence conservation among the sites and the lack of dyad symmetry within each site. The latter feature ensures that the sites operate to stop forks moving in one direction only.
The newly synthesised DNA strand is unmethylated and forms one part of a hemimethylated duplex. For this reason, the products of chromosome replication are chemically distinct from the template duplex until a full methylation of the newly synthesised strand has taken place. DNA adenine methyltransferase, Dam, methylates DNA at 5′‐GATC‐3′ sites and there are 11 of these sites in oriC ( Figure 1.2). The SeqA protein binds to these sites while they are still in their hemimethylated form, sequestering the origin and excluding DnaA (Han et al. 2003; Slater et al. 1995; von Freiesleben et al. 1994). The sequestered state persists for about one third of the cell cycle when it is relieved by dissociation of SeqA and methylation of the 5′‐GATC‐3′ sites by Dam (Kang, S. et al. 1999; Lu et al. 1994). SeqA also interferes with expression of the dnaA gene, reducing the levels of the DnaA protein available for binding to oriC (Campbell and Kleckner 1990). In addition, SeqA contributes to processes that ensure proper segregation of the chromosome copies at cell division (Helgesen et al. 2015; Waldminghaus and Skarstad 2009). It is interesting to note that both hemimethylated oriC and SeqA have been shown to associate with the cell envelope (Ogden et al. 1988; Slater et al. 1995), perhaps indicating a role for the complex in the positioning of oriC in the cell.
1.6 Replication Produces Physically Connected Products
The converging replication forks moving along the chromosome will create a topological problem as they approach one another in the Ter region. As chromosome replication comes to an end, the products that it generates will emerge as intertwined DNA duplexes. This physical linkage must be resolved if it is not to impede chromosome segregation. Furthermore, if homologous recombination occurs between the sister chromosomes it may create a chromosome dimer. This process is made more likely by oxidative damage to DNA, as occurs in mutants defective in the Fur iron regulatory protein (Steiner and Kuempel 1998). The dimers arise from RecBCD‐ and RecFOR‐mediated events with roughly equal frequency (Barre et al. 2001). Once formed, this dimer must be resolved at or before cell division or an anucleate cell will be created (see Section 1.8).
1.7 Decatenating the Sister Chromosomes
Bidirectional replication produces two interlinked copies of the chromosome and these must be decatenated before they can be segregated at cell division. Decatenation of fully intact duplexes is an intermolecular event that is catalysed by type II topoisomerases. Topoisomerase IV is the most efficient decatenase in such cases (Adams et al. 1992; Espéli et al. 2003; Zechiedrich and Cozzarelli 1995), although DNA gyrase can provide this function too (Steck and Drlica 1984). Type I topoisomerases can also supply a decatenase function, but in this case one of the DNA strands in at least one of the DNA duplexes must be nicked, with the topoisomerase providing the break in the intact strand of the same duplex to permit passage of the intact duplex through the gap (DiGate and Marians 1988).
1.8 Resolving Chromosome Dimers
The creation of chromosome dimers by homologous recombination between sister chromosomes threatens to interfere with chromosome segregation at cell division. The dimers are resolved by site‐specific recombination at the dif sites within the terminus regions of the cointegrated chromosomes. These sites are arranged as directly repeated sequences in the dimer, albeit 4.6 million base pairs apart, and dif synapsis, strand exchange, and resolution separate the chromosome copies as monomeric molecules ( Figure 1.8). The XerC and XerD tyrosine integrases act sequentially at dif to catalyse the reaction (Lesterlin et al. 2004; Sherratt et al. 1995); XerC creates the Holliday junction but this will collapse to substrate unless XerD completes the reaction (Barre et al. 2000; Hallet et al. 1999; Recchia and Sherratt 1999). The FtsK cell division protein triggers XerC/D‐mediated dimer resolution at dif (Steiner et al. 1999). FtsK is located at the cell division septum at the mid‐cell where it uses ATP hydrolysis to activate chromosome dimer resolution. Its location at the septum is dependent on its interaction with the FtsZ septum ring protein (Dorazi and Dewar 2000a; Yu et al. 1998; Wang and Lutkenhaus 1998). FtsK moves DNA within the cell, assists sister chromosome synapsis and reduces DNA entanglement/catenation of sister chromosomes (Sherratt et al. 2004). FtsK Orienting Polar Sequences (KOPS) located near the dif site in the Ter region of the chromosome contribute to the directional loading of the FtsK translocase, allowing it to segregate the daughter chromosome by driving a copy into each daughter cell (Bigot et al. 2006, 2007; Sivanathan et al. 2009; Stouf et al. 2013). The dif site and the wider Ter region of the chromosome remain in close association with the septum/nascent septum throughout the cell cycle (Niki et al. 2000).
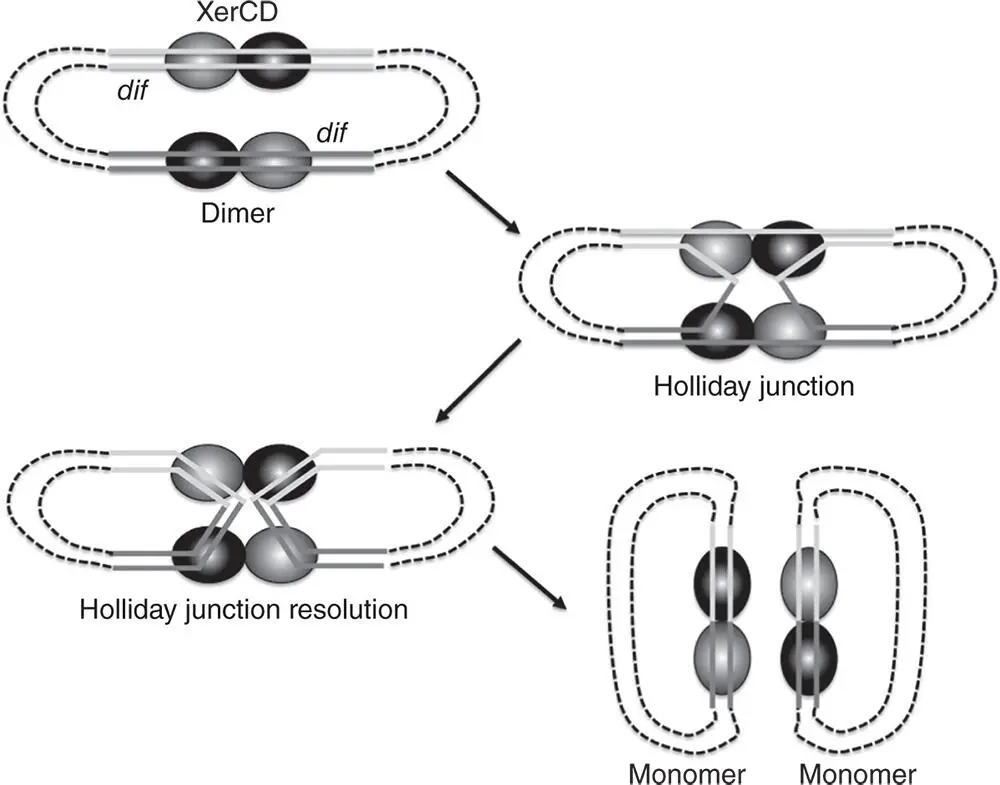
Figure 1.8Resolution of a chromosome dimer by XerCD‐mediated recombination at dif . Chromosome dimerisation can occur as a result of recombination‐mediated DNA repair during replication. The dimers are resolved by site‐specific recombination at two directly repeated dif sites located in the Ter region. The reaction is catalysed by the XerCD tyrosine integrases, generating monomeric chromosomes that can be segregated into the two daughter cells at cell division.
It is not in the interest of the bacterium to have chromosome dimers arising frequently because this poses a risk that either anucleate cells will arise or that the chromosome may be damaged by the division septum as the cell attempts to segregate a dimeric chromosome. RecA‐dependent homologous recombination events, whether they arise via the RecBCD or RecFOR pathways ( Section 1.6), generate Holliday junctions that must be resolved by the Ruv resolvasome‐based system. There is a bias in this process in favour of Ruv‐mediated resolution that does not involve crossover, and therefore the creation of chromosome dimers (Cromie and Leach 2000; van Gool et al. 1999). This bias reduces the frequency at which chromosome dimers, and the associated threat to the wellbeing of the daughter cell genomes, arise.
Читать дальше