1 ...8 9 10 12 13 14 ...29 The cis‐ acting parS centromere‐like sequences, and the genes that encode the ParA and ParB proteins, typically are found close to the oriC region of those bacterial chromosomes that harbour these systems (Livny et al. 2007; Reyes‐Lamothe et al. 2012). This centromere positioning ensures that the first part of the chromosome to be duplicated is going to be delivered to the appropriate cellular site for the orientation of the segregation of the rest of the chromosome. Positioning varies and can be at mid‐cell or quarter‐cell (Fogel and Waldor 2005; Webb et al. 1997); in the case of chromosomes with oriC tethering at the poles of rod‐shaped cells, it will be at the cell pole (Bowman et al. 2008; Fogel and Waldor 2006; Harms et al. 2013) ( Figure 1.11). Dimeric ParB is in excess compared with its multiple 16‐bp parS binding sites and it seems that ParB can spread beyond parS, perhaps by a bridging mechanism that has the effect of folding the centromere region into a tightly organised complex (Graham et al. 2014; Sanchez et al. 2013).
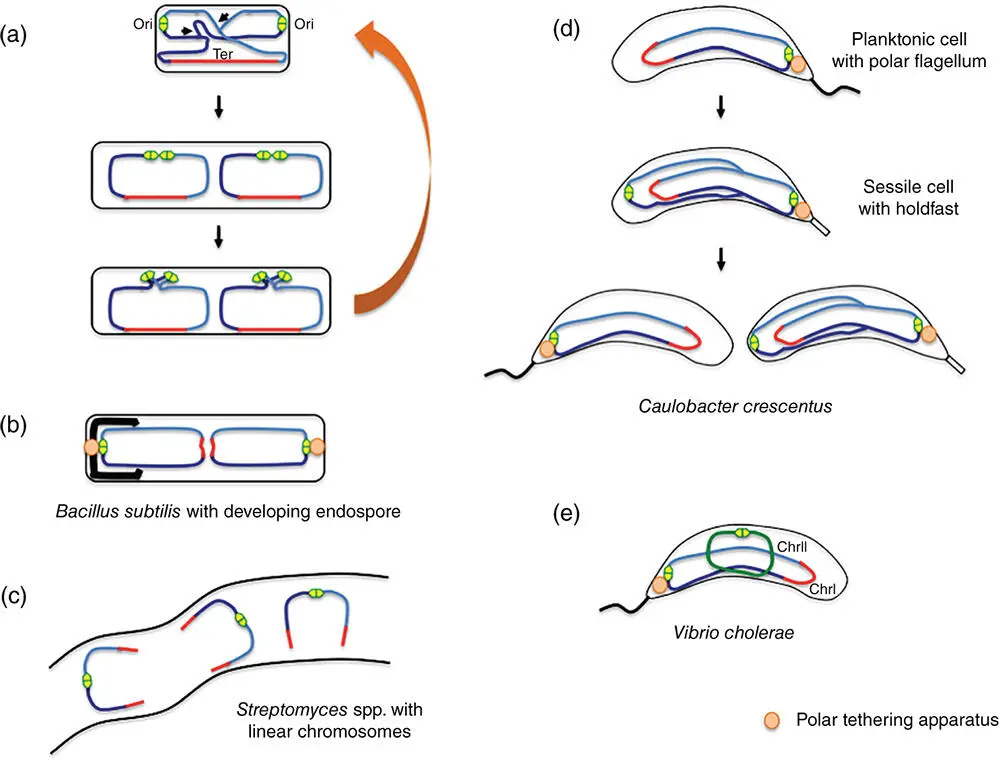
Figure 1.11Spatiotemporal dispositions of chromosomes in model bacteria other than E. coli . (a) In vegetative cells of Bacillus subtilis , the origins of chromosome replication are at mid cell in newborn cells before moving to the poles. The origins are at quarter cell positions at the onset of cell division. (b) In sporulating cells, the origins are tethered at the poles as one chromosome copy (left) is segregated into the developing fore‐spore. (c) Streptomyces species have linear chromosomes with the origin of replication at the midsection. (d) Caulobacter crescentus has two cell types: planktonic cells with a polar flagellum and sessile cells that adhere to the substratum via a polar holdfast. The origin of chromosome replication is tethered to the pole where the flagellum/holdfast is located. (e) Vibrio cholerae has two chromosomes. ChrI is the primary chromosome and its origin of replication is tethered to the cell pole where the single flagellum is located. The second chromosome, or chromid (ChrII), is plasmid‐like and synchronises its replication with that of ChrI.
The ParA protein drives bidirectional segregation of the parS‐ ParB complexes in an ATP‐dependent manner. It can form filaments, and these have been proposed to be a factor in chromosome segregation (Bouet et al. 2007; Hui et al. 2010; Ptacin et al. 2010). However, it is also possible that ParA‐driven chromosome segregation works by a diffusion‐ratchet‐type mechanism that has been described for its plasmid‐encoded counterparts (Hwang et al. 2013; Vecchiarelli et al. 2014) or a trans ‐nucleoid relay mechanism (Lim et al. 2014).
Ter and associated genetic loci replicate at mid‐cell and cohere for an extended period as a result of the matS‐ MatP nucleoprotein complex (Dupaigne et al. 2012; Reyes‐Lamothe et al. 2008; Mercier et al. 2008; Stouf et al. 2013; Wu et al. 2019). The C‐terminal domain of MatP interacts with the ZapB protein in the cell division apparatus and this interaction contributes to the extended cohesion of the Ter domain copies (Dupaigne et al. 2012; Espéli et al. 2012). MatP is displaced from matS by the action of the FtsK motor (or SpoIIIE in B. subtilis ) as it drives the Ter domain copies into the nascent daughter cells (Deghorain et al. 2011; Graham et al. 2010; Marquis et al. 2008; Massey et al. 2006; Sherratt et al. 2010). FtsK uses its dif‐ oriented KOPS repeats to bind and to guide this process; the XerCD recombination dif site is the final component of the chromosome to be segregated (Stouf et al. 2013). The formation of FtsK hexamers, triggered by the onset of cell division, is a critical step in FtsK's own activation (Bisicchia et al. 2013) and its activation of the XerCD recombination apparatus (Zawadzki et al. 2013), illustrating the exquisite integration of the Ter segregation and the chromosome dimer resolution systems.
1.10 Polar Tethering of Chromosome Origins
Anchoring the origin of replication to one pole of the cell is likely to assist in reinforcement of the ori‐Ter orientation of the chromosome seen along the long axis of rod shaped cells and in ensuring that daughter cells receive an entire chromosomal copy at cell division (Badrinarayanan et al. 2015) ( Figure 1.11). The PopZ protein fulfils this role in C. crescentus by forming a matrix at the pole and interacting with the ParB‐ parS complex at oriC (Bowman et al. 2008; Ebersbach et al. 2008). Displacement of parS to a different chromosome site interferes with this arrangement: while parS continues to be located at the pole oriC , from which parS is now disconnected, lies elsewhere in the cell (Umbarger et al. 2011).
The cytoplasmic protein HubP connects the origin of replication of ChrI to the cell pole in V. cholerae . The connection is made between HubP and the ParAI‐ParBI‐ parS complex. In addition to its membrane location, the HubP protein is connected to the cell wall through a peptidoglycan‐binding LysM motif, a feature that is required for its polar localisation (Yamaichi et al. 2012).
Polar attachment of the chromosome occurs in B. subtilis at the onset of sporulation. The RacA protein interacts with the DivIVA membrane protein that is located at the cell pole (Ben‐Yehuda et al. 2003; Lenarcic et al. 2009; Oliva et al. 2010; Ramamurthi and Losick 2009; Wu and Errington 2003). RacA also binds to ram (RacA binding motifs) that are found in 25 copies at oriC (Ben‐Yehuda et al. 2005). In the absence of RacA or DivIVA, sporulating bacteria fail to position the chromosome correctly and have the oriC at mid‐cell. This misplacement leads to the production of prespore compartments without chromosomes (Ben‐Yehuda et al. 2003). B. subtilis cells do not have their chromosomes attached to the cell pole during vegetative growth, although their origins occupy positions that alternate between pole‐proximal and at quarter‐cell, arrangements that require the cytoplasmic SMC complex (Wang, X., et al. 2014), just as the MukBEF equivalent in E. coli is required for that organism's chromosome to exhibit its customary ori‐ Ter orientation during rapid growth (Danilova et al. 2007).
1.11 Some Bacterial Chromosomes Are Linear
Most of the literature on bacterial chromosomes describes work with covalently closed, circular molecules. On the face of it, chromosome circularity is not essential for survival: work with E. coli has shown that linearisation of its circular chromosome through a phage‐mediated process that leaves the ends closed by DNA hairpins does not interfere significantly with the life of the bacterium (Cui et al. 2007). Going in the other direction, the linear chromosome of Streptomyces lividans can be circularised without killing the microbe, although its genetic instability increases (Volff et al. 1997).
Some organisms have linear chromosomes naturally. For example, Borrelia burgdorferi , the spirochete and causative agent of Lyme disease, has a complex genome consisting of a linear chromosome and 23 plasmids, some of which are circular while others are linear (Chaconas and Kobryn 2010). Essential metabolic functions are encoded by the plasmids, so these are parts of the core genome and not simply ancillary components. The ends of the linear DNA molecules are closed covalently by hairpin telomere‐like structures (Barbour and Garon 1987). Such structures are not found widely in bacteria; other examples have been reported in the plant pathogen Agrobacterium tumefaciens and in some phage (Chaconas and Kobryn 2010; Slater et al. 2013). Telomere resolvases, enzymes that are related to the integrase family of tyrosine site‐specific recombinases, promote fusions between the linear replicons at their telomeres, driving genome evolution (Huang et al. 2017). Replication of linear replicons in Borrelia spp. is thought to occur bidirectionally from a central origin, producing a double‐stranded dimeric circle that is resolved by the telomere resolvase (ResT in B. burgdorferi ) to produce two linear molecules with closed telomeres at their ends. Positive DNA supercoiling, probably arising from the local overwinding of the DNA during replication, assists telomere resolution (Bankhead et al. 2006). Although the role of DNA supercoiling, positive or negative, in linear replicons has not been studied comprehensively, there is some evidence that it is a factor in setting the level of transcription of promoters found on linear plasmids when those replicons are artificially circularised. This has led to the proposal that linear replicons may avoid instability caused by topological changes in circular molecules (Beaurepaire and Chaconas 2007). Streptomyces spp. also have linear chromosomes and linear plasmids, and intra‐replicon interactions mediated by ‘terminal proteins’ that are covalently bound to the telomeres allow the creation of negatively supercoiled DNA circles from the linear replicons (Tsai et al. 2011). These negative supercoils are relaxed by DNA topoisomerase I, which is a component of the telomere complex in Streptomyces . It has been proposed that negative supercoiling is likely to be important for both DNA replication and for transcription, especially of genes located close to the telomeres (Tsai et al. 2011).
Читать дальше