1.12 Some Bacteria Have More than One Chromosome
Among bacteria that have more than one chromosome are the well‐studied organisms A. tumefaciens (Allardet‐Servent et al. 1993), Brucella spp. (Jumas‐Bilak et al. 1998), Rhodobacter sphaeroides spp. (Choudhary et al. 2007; Suwanto and Kaplan 1989), and Vibrio spp. (Val et al. 2014). Of the organisms listed here, A. tumefaciens , has one circular and one linear chromosome; the others have two circular chromosomes. Paracoccus denitrificans is a bacterium that has three chromosomes (Winterstein and Ludwig 1998).
Are all of the chromosomes in a multi‐chromosome genome ‘equal’? It appears that one chromosome is usually the primary replicon, with the other being relegated to secondary chromosome status. For example, the two chromosomes of the pathogen V. cholerae are designated chromosomes, ChrI and ChrII, with ChrI having the majority of the metabolically important and virulence‐associated genes. ChrII does harbour essential genes, so its designation as a second chromosome is justified, despite its having plasmid‐like features. For example, the origin of replication of ChrII shows structural features that are similar to those of plasmids, which is consistent with the secondary chromosome having evolved from an ancestral plasmid (Orlova et al. 2017). The plasmid‐like nature of ChrII is also emphasised by its encoding plasmid RK2‐like toxin systems that ensure post‐segregation killing of those V. cholerae cells that lose ChrII (Yuan et al. 2011). The replication initiation system of ChrI resembles that of E. coli : initiation of replication of each chromosome is independent of, but coordinated with, that of the other (Duigou et al. 2006). ChrII begins replicating later in the cell cycle than the larger ChrI, but both finish together. This has been interpreted as a mechanism that compensates for the differences in size of the two molecules and the need to end replication simultaneously so that the chromosomes can be segregated together and that any dimers can be resolved simultaneously (Rasmussen et al. 2007). Further investigation has revealed the generality of coordinated termination of replication in members of the Vibrionaceae with two chromosomes (Kemter et al. 2018). ChrI and ChrII each possess their own ParAB‐ parS systems and use these for efficient segregation of chromosome copies at cell division (Fogel and Waldor 2005; Yamaichi et al. 2007, 2011).
In many bacteria, autonomously replicating and segregating genetic elements called plasmids accompany the chromosome in the cell. Like most bacterial chromosomes, plasmids are usually covalently closed, circular DNA molecules, but this is not always the case: some are linear. Certain plasmids are categorised as additional chromosomes (or ‘chromids’) due to their size, their carriage of genes normally found on bona fide chromosomes, their unitary copy number, and/or the coordination of their replication and segregation with the main chromosome (Barloy‐Hubler and Jebbar 2008; Fournes et al. 2018). Other very big plasmids are called ‘mega‐plasmids’ and can encode functions required for symbiosis or virulence (Schwartz 2008). In general, plasmids carry genes that are useful rather than essential, so their loss is not usually fatal to the cell; in contrast, loss of the chromosome is fatal.
Plasmids came to attention due to their involvement in bacterial sex (the Fertility, or F factor) and when it was discovered that they carried genes for resistance to antimicrobial agents, including antibiotics (R factors). Investigations of these phenomena led to the discovery of plasmid conjugation and the existence of other mobile genetic elements such as transposons and integrons. Plasmid studies revealed a wealth of information about plasmid replication processes, segregation systems, and copy number control mechanisms. This field also provided cloning vectors to support the emergence of the recombinant DNA technology that, in part, underpins biotechnology. Plasmid research has provided important insights into gene regulation mechanisms, including the provision of early examples of the regulatory roles of small RNA molecules.
The term plasmid was introduced in the mid‐twentieth century to describe self‐replicating extrachromosomal DNA elements (Lederberg 1952). Self‐transmissible plasmids are important contributors to HGT; indeed, some plasmids can even participate in DNA transfer between different domains of life (Suzuki et al. 2008; Zambryski et al. 1989). Limits to the host range of a plasmid include barriers to mating bridge establishment, to successful DNA transfer, and to successful plasmid replication (Jain and Srivastava 2013). Plasmid size is a poor predictor of host range: while many broad‐host‐range plasmids are large, many others are quite small. For example, pBC1 can function in both Gram‐negative and Gram‐positive bacteria, yet it is only 1.6 kb in size (De Rossi et al. 1992). At the other end of the scale, the intensively studied RK2/RP4 plasmid group is in the 60‐kb‐size range but is confined to Gram‐negative hosts, albeit a wide selection of these (Thomas et al. 1982). It is an advantage to have several origins of plasmid replication as this improves the chances of being able to replicate in a given host. However, the presence of multiple origins is not in itself a reliable indicator of broad host range: the F plasmid has a narrow range yet it has three origins of replication. Instead, it is the structure of the origin(s) that seems to be important in determining host range. Plasmids with a minimum dependence on host‐encoded factors for replication are likely to have a broad host range; for example, RSF1010 from incompatibility group Q (IncQ) uses a strand‐displacement mode of replication that relies on no host‐encoded factors for the initiation of DNA synthesis (Loftie‐Eaton and Rawlings 2012).
The RK2 plasmid (IncP) has a broad host range and can replicate in E. coli or Pseudomonas aeruginosa (Shah et al. 1995). It can replicate in E. coli using just a minimal origin, oriV (‘vegetative’ origin), containing five iterons (directly repeated sequences) and four binding sites for DnaA (Doran et al. 1999) ( Figure 1.12). In P. aeruginosa , RK2 needs an additional three iterons but can dispense with the DnaA boxes (Schmidhauser et al. 1983). RK2 is also capable of replicating without DnaA when in C. crescentus (Wegrzyn et al. 2013).
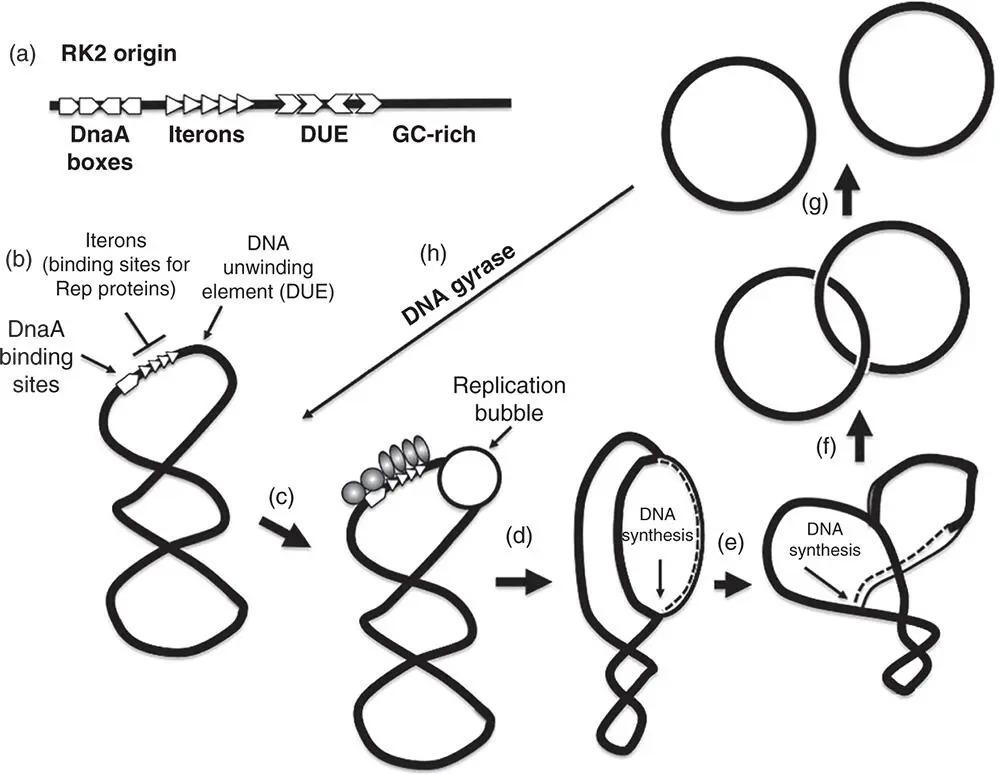
Figure 1.12Theta model of plasmid replication. (a) The structure of the origin of replication in the broad‐host‐range, single‐copy, IncP plasmid RK2, showing the relative positions and numbers of the DnaA boxes, the iterons (binding sites for the replication protein, TrfA), the AT‐rich DNA unwinding element (DUE), and the adjacent GC‐rich sequence. (b) A replication cycle is shown for an idealised plasmid using theta replication. Replication begins with the binding of the replication protein to the iterons and the recruitment of the host‐encoded DnaA to the DnaA boxes. (c) The DNA in the DUE becomes single‐stranded, creating a replication bubble to which host‐encoded replication proteins are recruited. (d, e) Depending on the plasmid, DNA synthesis proceeds either uni‐ (e.g. ColE1) or bidirectionally (e.g. RK2). (f) The products of plasmid replication are catenated, double‐stranded circles and these are unlinked by Topo IV. (g, h) The unlinked plasmids are negatively supercoiled by DNA gyrase, recreating the substrate for another round of replication.
Читать дальше