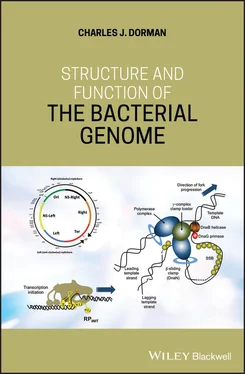
Figure 1.2Structure of oriC on the E. coli chromosome. The ATP‐dependent DnaA protein binds to sites throughout oriC and oligomerises in the DnaA Oligomerisation Region (DOR), driving DNA unwinding at the A+T‐rich DNA Unwinding Element (DUE). Single‐stranded T‐rich DNA in DUE binds to the DnaA oligomers at DOR. High‐affinity sites bind DnaA‐ATP or DnaA‐ADP; low affinity sites bind just DnaA‐ATP. Binding sites for the NAPs FIS and IHF are also shown: FIS and IHF modulate the process of replication initiation negatively and positively, respectively. The Dam methylase methylates oriC at several 5′‐GATC‐3′ sites (indicated by vertical arrows): hemimethylated sites bind SeqA, excluding DnaA and preventing untimely re‐initiation of chromosome replication.
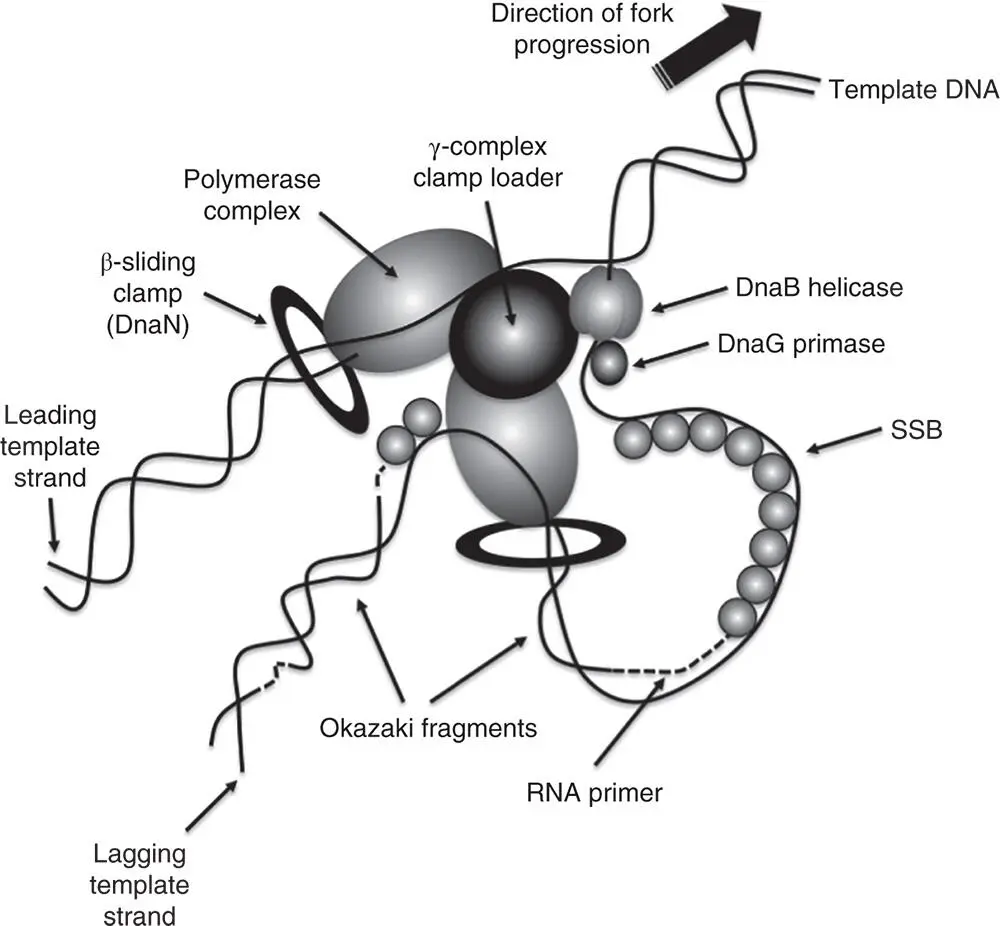
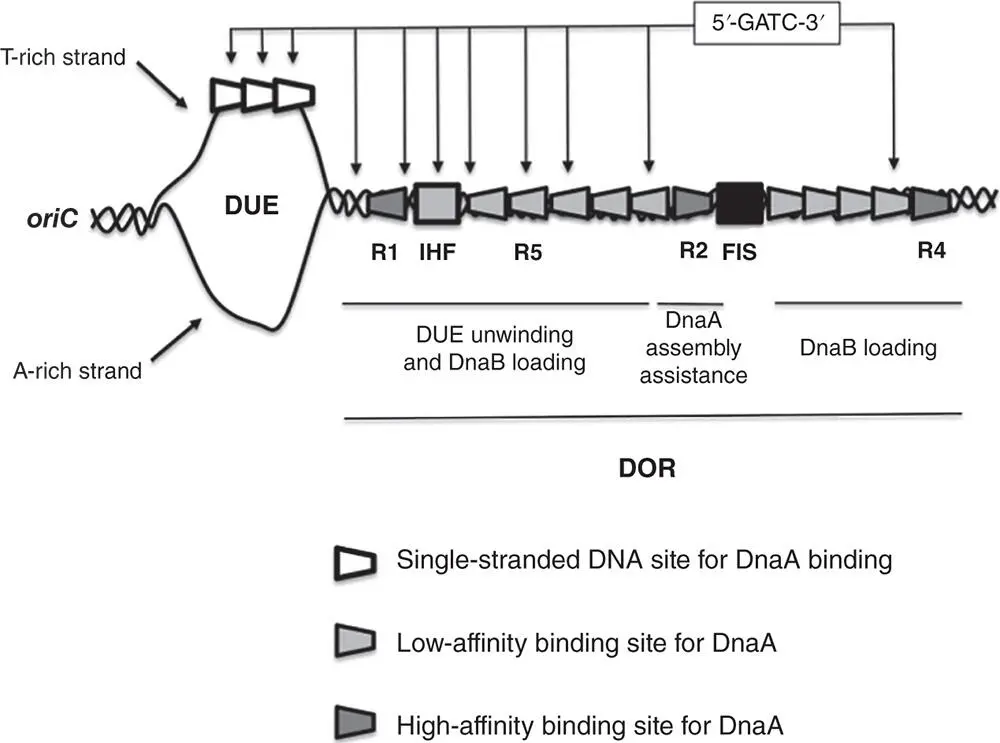
Figure 1.3Structure of the E. coli replisome in chromosome replication. The replisome is made up of the two cores of DNA Polymerase III, a gamma (γ) complex (or clamp loader) and the beta clamp together with a hexameric helicase, the DnaG primase, and the single‐stranded binding protein, SSB. The DnaB helicase uses energy from ATP hydrolysis to translocate along the lagging strand, unwinding the DNA duplex. The two Polymerase III cores, linked by the tau subunits ( Figure 1.4), are each dedicated to coordinated and simultaneous replication of the leading and lagging template strands of the replication fork. The ring‐like beta (β) clamp (DnaN), or processivity factor, encircles DNA and is attached to the replisome via the alpha (α) subunit. The β clamp stabilises the moving replication machine on its template, allowing it to operate with a high degree of processivity. A single‐stranded DNA bubble is formed by the unwinding action of the replisome and SSB protein coats the ssDNA. The DnaG primase interacts with the helicase to generate RNA primers that are used to prime Okazaki fragment synthesis.
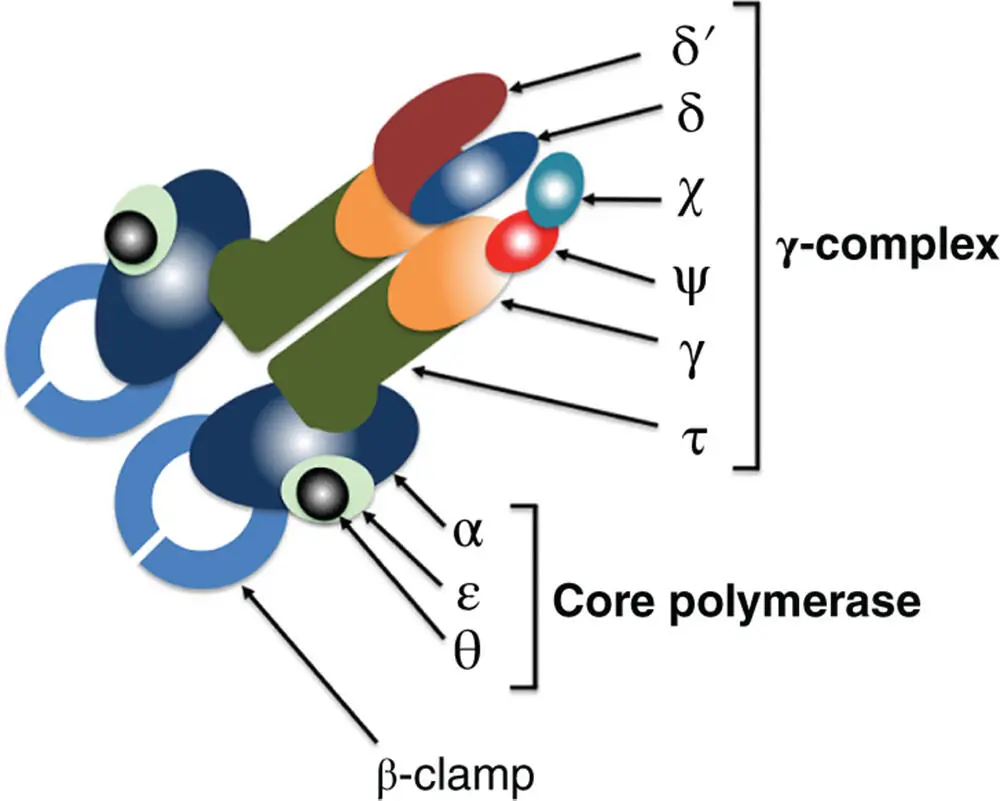
Figure 1.4The structure of the DNA polymerase III subassembly (Pol III*). The core is made up of the alpha (α), epsilon (ɛ), and theta (θ) subunits and the holoenzyme contains two cores. The tau (τ) subunit (two copies) links the cores together, ensuring simultaneous replication of the leading and lagging strands. The function of the cores is DNA synthesis on the leading and lagging strands ( Figure 1.3). The clamp loader (or gamma complex) is made up of the chi (χ), delta (δ), delta‐prime (δ′), gamma (γ), and psi (ψ) subunits. The gamma subunit loads the beta clamp onto the DNA that is primed for de novo DNA synthesis. The arrival of the beta (β) clamp (processivity factor) converts the Pol III* subassembly into the Pol III holoenzyme ( Figure 1.3).
In oriC of E. coli , the DnaA boxes are of variable affinity for the DnaA protein (Blaesing et al. 2000) ( Figure 1.2). Boxes with high affinity bind DnaA that is in a complex with either ATP or ADP, whereas weak boxes bind only DnaA that has bound ATP (Grimwade et al. 2007). Binding of DnaA to oriC is cooperative, with DnaA‐ATP that has bound to strong boxes facilitating the subsequent binding of DnaA‐ATP to the weaker sites, promoting the formation of the DnaA oligomer at the origin of replication (Miller et al. 2009; Kaur et al. 2014). The activity of DnaA may also be controlled by reversible acetylation at lysine residues: of the 13 lysine amino acids in DnaA, acetylation of residues K178 and K243 seems to be especially important in promoting the initiation of chromosome replication (Li et al. 2017; Zhang, Q., et al. 2016).
The availability of DnaA‐ATP is a rate‐limiting factor for the initiation of chromosome replication. A protein called Hda converts active DnaA‐ATP into inactive DnaA‐ADP through ATP hydrolysis (Kato and Katayama 2001). This conversion also requires DnaN, the DNA polymerase beta‐clamp, linking ATP hydrolysis to the elongation phase of DNA synthesis (Takata et al. 2004).
The nucleoid‐associated proteins (NAPs) Integration Host Factor (IHF) and the Factor for Inversion Stimulation (FIS) are DNA‐binding and ‐bending proteins that are thought to play important architectural roles at the origin of replication ( Figure 1.2) (Kasho et al. 2014; Ryan et al. 2004). IHF has a positive role at oriC where it binds to a specific DNA sequence, introducing a DNA bend that encourages DnaA binding and oligomer formation; it can also redistribute DnaA on supercoiled DNA (Grimwade et al. 2000). While some work has not found a major role for FIS in regulating events at oriC (Weigel et al. 2001) data from other investigations show that, in contrast to IHF, the role of FIS is inhibitory to DNA replication: when it binds to oriC it interferes with the binding of IHF and DnaA, blocking unwinding of the DUE sequence (Ryan et al. 2004).
The many 5′‐GATC‐3′ sites found throughout oriC ( Figure 1.2) are hemimethylated in the period immediately following the initiation of chromosome replication (Lu et al. 1994). The SeqA protein binds to these hemimethylated sites, preventing immediate and untimely re‐initiation of chromosome replication by DnaA: SeqA also downregulates the expression of the negatively autoregulated dnaA gene (Campbell and Kleckner 1990; Waldminghaus and Skarstad 2009). Dam‐mediated methylation of the 5′‐GATC‐3′ sites is inhibitory to SeqA binding and re‐admits DnaA to oriC (Lu et al. 1994).
E. coli uses clusters of DnaA binding sites that are located outside oriC to modulate the initiation of chromosome replication ( Figure 1.5). One of these is the 183‐bp datA site, located next to the vjeV gene on the E. coli chromosome. The datA site is made up of five high‐affinity DnaA binding sites (Kitagawa et al. 1996); datA also binds IHF (Nozaki et al. 2009). The interaction of IHF with datA occurs immediately after the initiation of chromosome replication and this facilitates the binding of DnaA‐ATP to datA (Nozaki et al. 2009). DnaA‐ATP bound to datA undergoes ATP hydrolysis, reducing the size of the pool of DnaA‐ATP that is available for binding to oriC (Ogawa et al. 2002). This IHF‐dependent process has a negative influence on the frequency with which chromosome replication is initiated at oriC (Kasho and Katayama 2012).
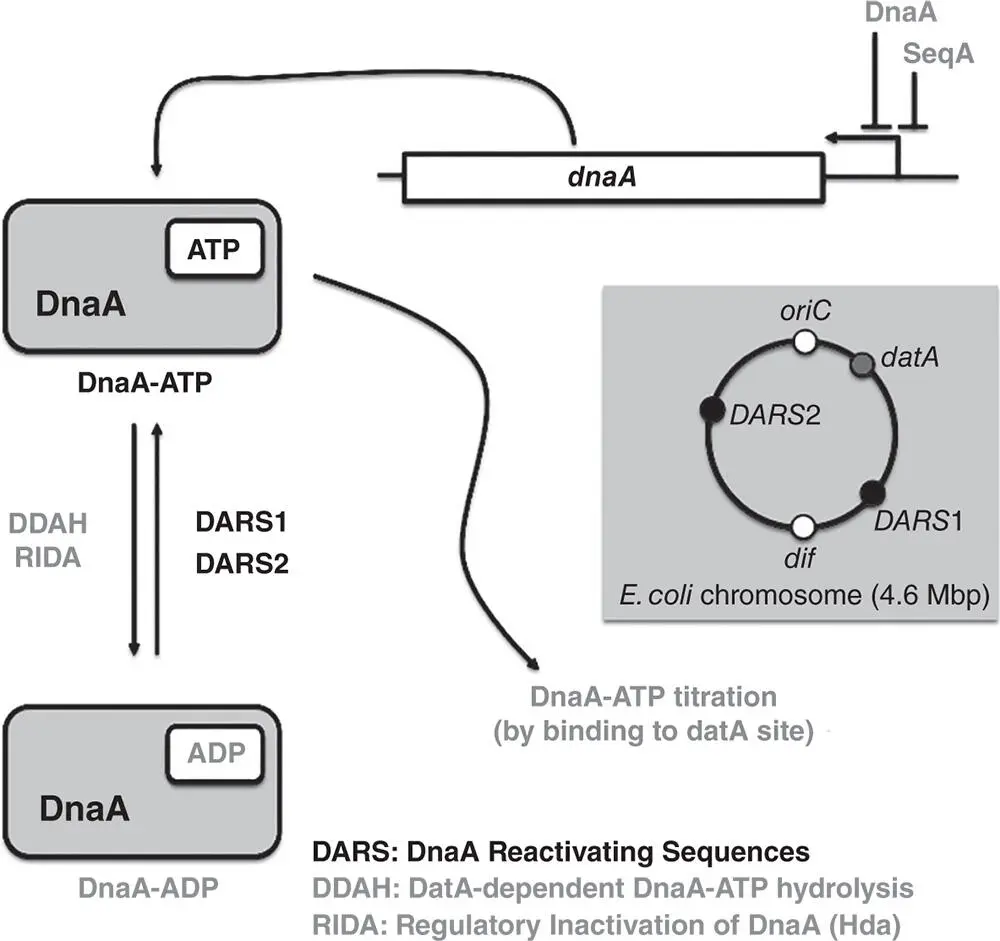
Figure 1.5The control of DnaA production and activity. The SeqA and DnaA proteins regulate expression of the dnaA gene negatively. DnaA‐ATP is generated at the DnaA Reactivating Sequences DARS1 and DARS2, and is converted to DnaA‐ADP by ATP hydrolysis (i) at the datA site stimulated by binding of IHF in a process called datA ‐dependent DnaA‐ATP Hydrolysis (DDAH) and (ii) by Regulatory Inactivation of DnaA (RIDA) in which the DnaA inhibitor protein Hda catalyses the hydrolysis of DnaA‐bound ATP to ADP, yielding DnaA‐ADP. Hda activation in RIDA follows interaction with the DNA polymerase clamp on newly synthesised DNA. The relative locations of datA (4.39 Mb), DARS1 (0.81 Mb) and DARS2 (2.97 Mb) with respect to the oriC and dif sites on the 4.6 Mb E. coli chromosome are shown (inset). Black lettering: generation of DnaA‐ATP, grey lettering: conversion of DnaA‐ATP to DnaA‐ADP.
Читать дальше