Jane Flint - Principles of Virology
Здесь есть возможность читать онлайн «Jane Flint - Principles of Virology» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Principles of Virology
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:3 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 60
- 1
- 2
- 3
- 4
- 5
Principles of Virology: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Principles of Virology»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
Volume I: Molecular Biology
Volume II: Pathogenesis and Control
Principles of Virology, Fifth Edition
Principles of Virology — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Principles of Virology», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
The crystal structures of the four types of nucleic acid polymerases reveal that the enzymes resemble a right hand consisting of a palm, fingers, and a thumb, with the active site of the enzyme located in the palm ( Fig. 6.4B). This shape supports the correct optimal arrangement of substrates and metal ions at the catalytic site and allows the dynamic changes needed during nucleic acid synthesis. The structures of RdRPs differ in detail from those of other polymerases, presumably to accommodate different templates and priming mechanisms. All nucleic acid polymerases have a similar core catalytic domain configuration and evolved from a common ancestor.
High-resolution structures of RdRPs and complexes with RNA in the process of catalysis have been determined for many (+) strand RNA, (–) strand RNA, and double-stranded RNA viruses. The RdRPs of picornaviruses and caliciviruses are the smallest known polymerases. Their structures are at the core of polymerases from larger RNA viruses, which typically contain additional domains that provide other replication-linked functions, such as methyltransferase, RNA capping, a platform for primer-independent initiation, and membrane anchoring.
The fingers and thumb subdomains of RdRPs show extensive interactions that encircle the active site and form a channel in which the template binds ( Fig. 6.4). The closed structure creates a nucleoside triphosphate (NTP) entry tunnel on one face of the enzyme and a template-binding site on the other. Residues within motif F, a conserved region unique to RdRPs ( Fig. 6.4), form the NTP entry tunnel, while motif G is in a loop that outlines the template entry channel. In contrast, structures of other polynucleotide polymerases resemble an open hand.
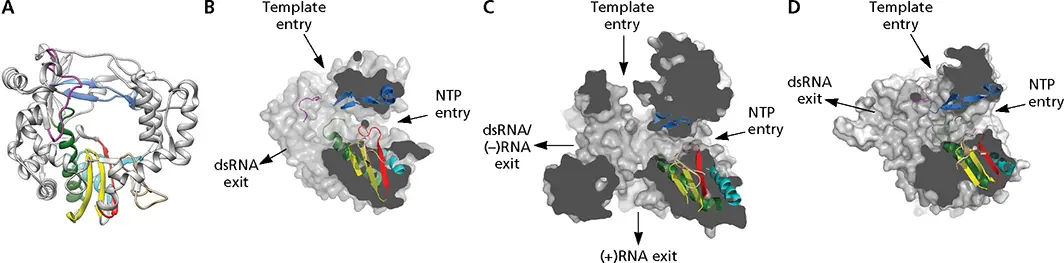
Figure 6.5 Structural elements of viral RNA-dependent RNA polymerase. (A)Ribbon representation of poliovirus 3D pol(PDB file 3OL6). Conserved motifs are colored: motif A, red; motif B, green; motif C, yellow; motif D, cyan; motif E, tan; motif F, blue; motif G, magenta. (B-D)Surface representations of three RdRP enzymes are cut to expose channels that are the entry and exit sites of substrates and reaction products. Motifs A to G are colored as in panel A. (B)Poliovirus RdRP (PDB, 3OLB); (C)reovirus λ3 RdRP (PDB, 1N35). (D)Bacteriophage phi6 RdRP (PDB, 1HI0). Images B–D courtesy of Núria Verdaguer, Molecular Biology Institute of Barcelona.
Three channels can be observed in the structures of RdRPs from (+) strand RNA and some double-stranded RNA viruses, which comprise the entry and exit paths for template and an NTP channel ( Fig. 6.5). In the polymerases of reovirus and (–) strand RNA viruses, N- and C-terminal extensions of the core enzyme form cage-like structures. In these enzymes, the buried active sites are connected to the exterior by four channels. The NTP and template entry channels lead to the catalytic site in the palm subdomain. The palm comprises a three-stranded antiparallel β-sheet that is surrounded by three α-helices and contains the four structural motifs in the order A-B-C-D. Motif A contains the consensus sequence DX 4-5C (where X is any amino acid), while motif C includes the amino acid triplet XDD, which is Gly-Asp-Asp in the RdRPs of most (+) strand RNA viruses. The two Asp residues of motif C and the conserved Asp238 of motif A form a cluster that coordinates the triphosphate moiety of the NTP substrate and the metal ions required for catalysis. Motif E, which is present in RNA-dependent but not in DNA-dependent polymerases, lies between the palm and thumb domains ( Fig. 6.5). It projects into the active site and helps to position the 3′ end of the RNA primer.
The thumb domains of picornavirus and calicivirus RdRPs are small, and as a consequence, a large central cleft is present on one side of the molecule. This cleft accommodates a protein primer during initiation, and the double-stranded RNA product during elongation. In contrast, the polymerases of flaviviruses have much larger thumb domains with elements that protrude into the template channel and provide priming platforms for de novo initiation (see below).
RdRPs preferentially incorporate NTPs rather than deoxyribonucleoside triphosphates (dNTPs). NTP recognition by poliovirus 3D polis regulated by Asp238 of motif A, which forms a hydrogen bond with the ribose 2′-OH ( Fig. 6.6). dNTPs are not bound because Asp238 cannot form a hydrogen bond with 2′-deoxyribose. An Asp is present at this position in all RdRPs. A Tyr at this position in RNA-dependent DNA polymerases (reverse transcriptases) is responsible for discriminating against NTPs and selecting dNTPs.
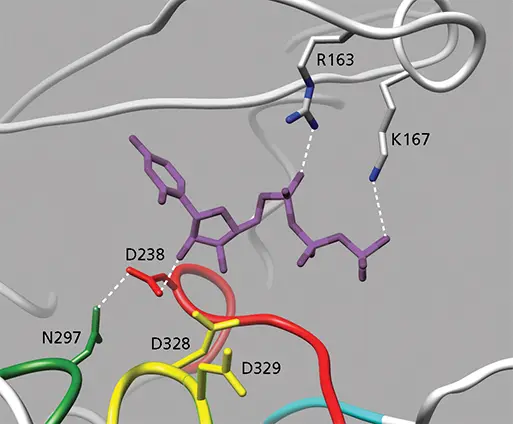
Figure 6.6 Structure of UTP bound to poliovirus 3D pol. The NTP bridges the fingers (top) and palm (bottom) domains. The base is stacked with Arg163 from the fingers. Hydrogen bonds are shown as dashed lines. The Asp238 of motif A, which is conserved in all RNA-dependent RNA polymerases, hydrogen bonds with the 2′-OH of the ribose moiety; this interaction discriminates NTPs from dNTPs. Asp328 and Asp329, which coordinate Mg 2+, are also labeled (PDB file 2IM2).
Mechanisms of RNA Synthesis
Initiation
As polymerases synthesize nucleic acid in a 5′ to 3′ direction, the nucleotidyl transfer reaction is initiated at the 3′ end of the template strand. The requirement for a primer for initiation of nucleic acid synthesis varies among the different classes of polymerases. Most DNA polymerases are primer-dependent enzymes, while DNA-dependent RNA polymerases initiate RNA synthesis de novo . Some RdRPs (e.g., those of flaviviruses and rhabdoviruses) can also initiate RNA synthesis de novo , while others require a primer ( Fig. 6.7). In these cases, RNA polymerization is initiated by a protein-linked primer (picornaviruses) or an oligonucleotide cleaved from the 5′ end of cellular pre-mRNA (influenza viruses).
De Novo Initiation
In this process, the first phosphodiester bond is made between the 3′-OH of the initiating NTP and the second NTP ( Fig. 6.7). In these cases, initiation takes place at the exact 3′ end of the template, except during replication of the genomes of some (−) strand RNA viruses, such as bunyaviruses and arenaviruses ( Fig. 6.7). Initiation begins at an internal C, and after extension of a few nucleotides, the daughter strand is shifted in the 3′ direction so that the 5′-terminal G residue is not base paired with the template strand. Because the daughter strand slips, this mechanism is called “prime and realign.”
De novo initiation
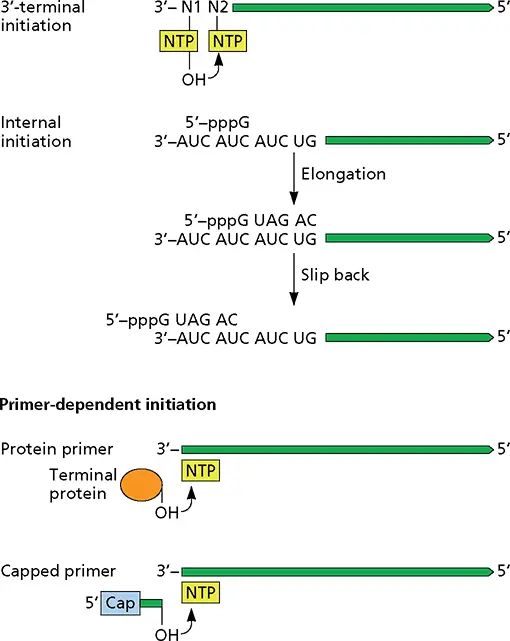
Figure 6.7 Mechanisms of initiation of RNA synthesis. De novo initiation may occur at the 3′ end of the viral RNA or from an internal base. When a primer is required, it may be a capped or protein-linked oligonucleotide.
Structural comparisons of viral RdRPs that catalyze de novo initiation reveal larger thumb subdomains with structural elements that fill most of the active-site cavity and provide a platform for initiating nucleotides. For example, the structure of the RdRP of hepatitis C virus indicates that a dinucleotide is synthesized by the polymerase using a β-loop insertion in the thumb domain as a “protein platform” in the active site ( Fig. 6.8). After the product reaches a certain length, the polymerase undergoes a conformational change that moves the priming platform out of the way and allows the newly synthesized complementary RNA to exit as the enzyme moves along the template strand.
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Principles of Virology»
Представляем Вашему вниманию похожие книги на «Principles of Virology» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Principles of Virology» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.











