Jane Flint - Principles of Virology
Здесь есть возможность читать онлайн «Jane Flint - Principles of Virology» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Principles of Virology
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:3 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 60
- 1
- 2
- 3
- 4
- 5
Principles of Virology: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Principles of Virology»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
Volume I: Molecular Biology
Volume II: Pathogenesis and Control
Principles of Virology, Fifth Edition
Principles of Virology — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Principles of Virology», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
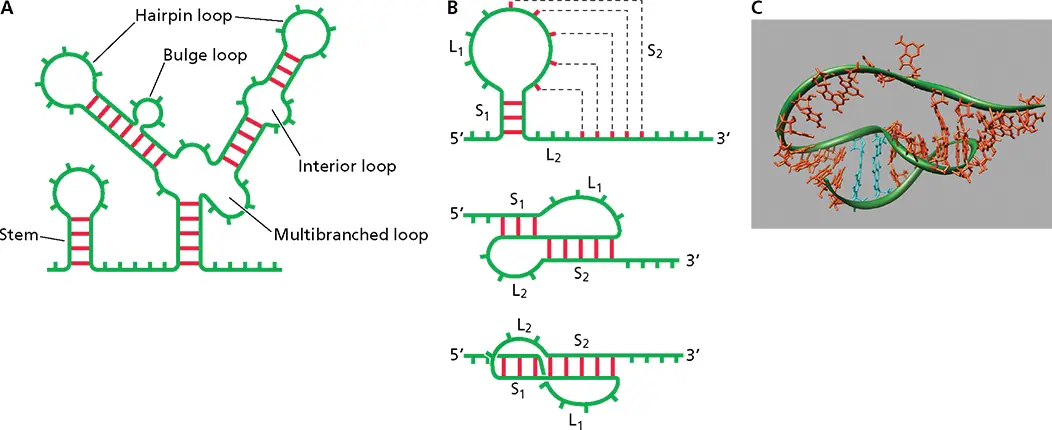
Figure 6.2 RNA secondary structure. (A)Schematic of different structural motifs in RNA. Red bars indicate base pairs; green bars indicate unpaired nucleotides. (B)Schematic of a pseudoknot. (Top) S tem 1 (S 1) is formed by base pairing in the stem-loop structure, and stem 2 (S 2) is formed by base pairing of nucleotides in the loop with nucleotides outside the loop. (Middle) A different view of the formation of stems S 1and S 2. (Bottom) Coaxial stacking of S 1and S 2resulting in a quasicontinuous double helix. (C)Structure of a pseudoknot as determined by X-ray crystallography. The sugar backbone is highlighted with a green tube. Stacking of the bases in the areas of S 1and S 2can be seen (PDB file 1L2x). Adapted from Pleij CW. 1990. Trends Biochem Sci 15:143–147, with permission.
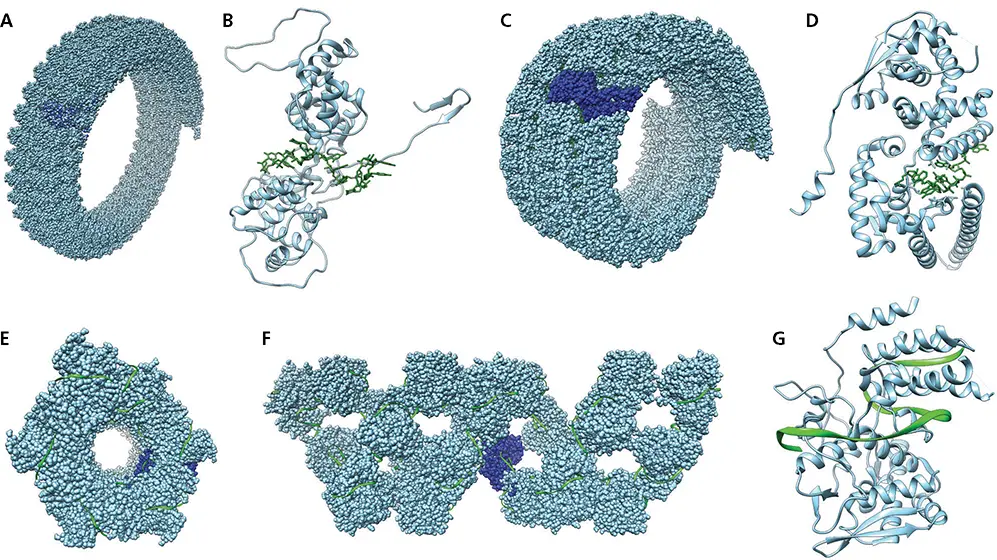
Figure 6.3 Structure of viral ribonucleoproteins. (A)Space-filling model of vesicular stomatitis virus partial helical filament formed by the nucleoprotein-RNA complex. Protein is colored blue and RNA green; a single nucleoprotein subunit is colored dark blue (PDB file 2WYY). (B)Ribbon diagram of vesicular stomatitis virus N protein monomer bound to RNA (colored green) (PDB file 2WYY). (C)Space-filling model of Ebolavirus partial helical filament formed by the nucleoprotein-RNA complex. Protein is colored blue and RNA green; a single nucleoprotein subunit is colored dark blue (PDB file 6NUT). (D)Ribbon diagram of Ebolavirus nucleoprotein (blue) monomer bound to RNA (green) (PDB file 6NUT). (E)Space-filling model of helical influenza virus NP bound to RNA, viewed from one end down the central axis. One NP monomer is colored dark blue (PDB file 4BBL). (F)Space-filling model of helical influenza virus NP bound to RNA, viewed from the side. One NP monomer is colored dark blue. (PDB file 4BBL). (G)Ribbon diagram of influenza virus nucleoprotein monomer (blue) bound to RNA (green) (PDB file 4BBL).
The initial discovery of a putative RdRP in poliovirus-infected cells was followed by attempts to purify the enzyme and show that it can copy viral RNA. Because polioviral genomic RNA contains a 3′ poly(A) sequence, polymerase activity was measured with a poly(A) template and an oligo(U) primer. A poly(U) polymerase was purified from infected cells and shown to copy polioviral genomic RNA in the presence of this primer. Poly(U) polymerase activity coincided with a single polypeptide, now known to be the polioviral RdRP 3D pol(see Appendix, Fig. 21, for a description of this nomenclature). Purified 3D polcannot copy polioviral genomic RNA in the absence of a primer.
Such assays for RdRP activity have been used to detect the presence of virus-specific enzymes in virus particles or in extracts of cells infected with a wide variety of RNA viruses. Amino acid sequence alignments can be used to identify viral proteins with motifs characteristic of RdRPs. These approaches were applied in identification of the L proteins of paramyxoviruses and bunyaviruses, the PB1 protein of influenza viruses, and the nsP4 protein of alphaviruses as candidate RdRPs. When the genes encoding these polymerases are expressed in cells, the proteins that are produced can copy viral RNA templates.
RNA-directed RNA synthesis obeys a set of universal rules. RNA synthesis is catalyzed by virus-encoded polymerases and initiates and terminates at specific sites in the template, but viral accessory proteins and even host cell proteins may also be required. Some RdRPs can initiate RNA synthesis de novo . Others require a primer with a free 3′-OH end to which nucleotides complementary to the template are added. Some RNA primers are protein linked, while others bear a 5′ cap structure (the cap structure is described in Chapter 8). A comparison of the structures and sequences of polynucleotide polymerases has led to the generality that all DNA and RNA polymerases catalyze synthesis by a mechanism that requires two metals ( Box 6.2). RNA is usually synthesized by template-directed, stepwise incorporation of ribodeoxynucleoside monophosphates (NMPs) into the 3′-OH end of the growing RNA chain, which undergoes elongationin the 5′ → 3′ direction.
Three-Dimensional Structures of RNA-Dependent RNA Polymerases
Viral RdRPs show the highest degree of conservation of all viral proteins and are the only proteins common to all RNA viruses. Seven conserved sequence motifs (A to G) that contain amino acid residues crucial for enzymatic function have been identified in all RdRPs ( Fig. 6.4). Some of the conserved sequence motifs are shared among all four classes of nucleic acid polymerases ( Fig. 6.4).
BOX 6.2
BACKGROUND
Two-metal mechanism of catalysis by polymerases
All polynucleotide polymerases are thought to catalyze synthesis by a two-metal mechanism that requires two conserved aspartic acid residues (illustrated in the figure for a DNA polymerase). The carboxylate groups of these amino acids coordinate two divalent metal ions, shown as Mg 2+in the figure. One metal ion promotes deprotonation of the 3′-OH group of the nascent strand, and the other ion stabilizes the transition state at the α-phosphate of the NTP substrate and facilitates the release of pyrophosphate (PP i).
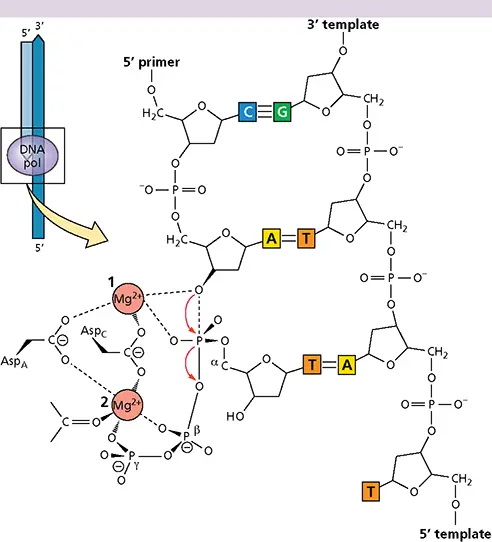
Two-metal mechanism of DNA polymerase catalysis.Red arrows indicate the net movement of electrons.
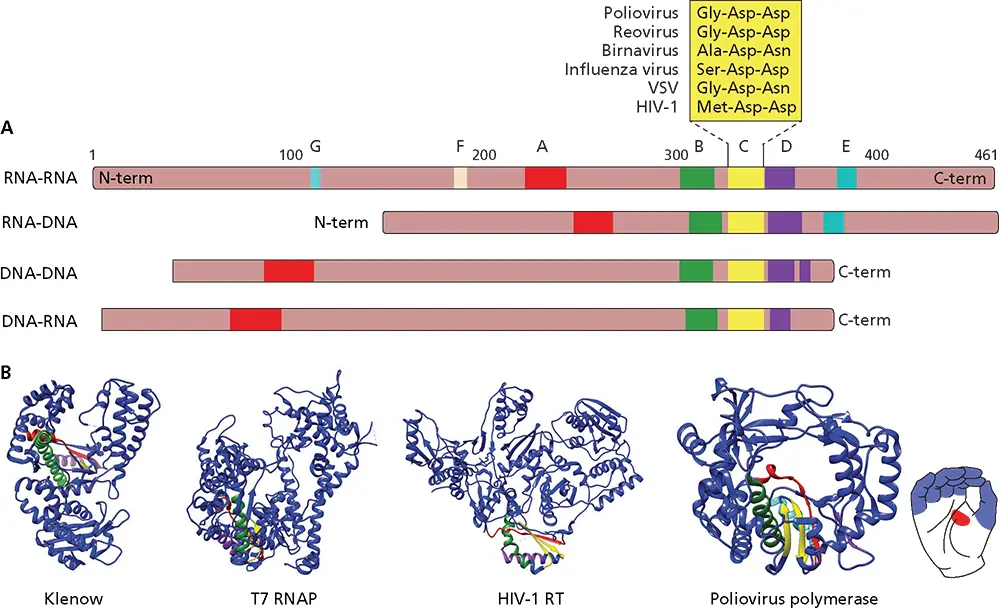
Figure 6.4 Protein domain alignments for the four categories of nucleic acid polymerases. (A)Schematic diagrams of polymerases. Numbers at the top are from the poliovirus 3D polamino acid sequence. Sequence and structure motifs in each polymerase category are colored. Motif F is found only in RNA-dependent RNA polymerases. The Asp-Asp sequence of motif C is also conserved in RNA-dependent DNA polymerases of retroviruses and in RNA polymerases of double-stranded RNA and segmented (−) strand viruses. The RNA polymerases of nonsegmented (−) strand viruses contain Gly-Asp-Asn instead of Gly-Asp-Asp. Mutational studies have shown that this sequence in the RdRP (L protein) of vesicular stomatitis virus (VSV) is essential for RNA synthesis. The RdRP of birnavirus, an insect virus with a double-stranded RNA genome, has Ala-Asp-Asn instead of Gly-Asp-Asp. (B)Representative structures of each of the four types of nucleic acid polymerases. Ribbon diagrams of the polymerase domain of the large (Klenow) fragment of Escherichia coli DNA polymerase I, a DNA-dependent DNA polymerase; T7 RNA polymerase (T7 RNAP), a DNA-dependent RNA polymerase; human immunodeficiency virus type 1 reverse transcriptase (HIV-1 RT), an RNA-dependent DNA polymerase; and polioviral 3D pol, an RNA-dependent RNA polymerase. The thumb domain is at the right, and the fingers domain is at the left. The conserved structure/sequence motifs A, B, C, D, and E are colored red, green, yellow, purple, and cyan, respectively (PDB files 1QSL, 1S77, 4B3O, and 3OL6).
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Principles of Virology»
Представляем Вашему вниманию похожие книги на «Principles of Virology» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Principles of Virology» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.











