Search for resistance in wild relatives of tomato
As a consequence of inbreeding during tomato domestication, the genetic diversity in cultivated tomato is now very narrow. However, large variation is present and exploitable in the wild Solanum species. Thus, the first step was to find wild tomato accessions with resistance to tomato powdery mildew.
At the Tomato Genetics Resource Center in Davis, California (TGRC, http://tgrc.ucdavis.edu) and Botanical and Experimental Garden ( http://www.bgard.science.ru.nl) in the Netherlands, thousands of accessions of the wild Solanum species have been collected and maintained. From these collections, we have selected and tested some Solanum species with tomato powdery mildew. As expected, many wild accessions showed resistance ( Figure B5.1).
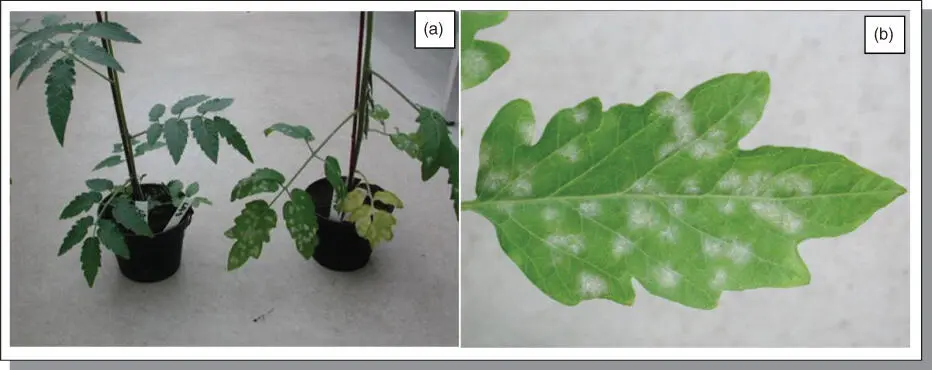
Figure B5.1Tomato plants inoculated with tomato powdery mildew. (a) The left plant is from tomato wild species Solanum pervianum LA2172, showing no powdery mildew infection; the right plant is from S. lycoerpsicum cv. Moneymaker (MM), showing fungal clonies growing on infected leaves. (b) A closer look at the colonization of tomato powdery mildew ( Oidium neolycopersici ) growing on the upper‐side of MM leaf. Pictures were taken 15 days post inoculation.
Inheritance of the resistance
Monogenic resistance is most exploited in tomato breeding programs. Modern tomato cultivars may harbor resistances to more than 10 pathogens. Thus, the second step is to study the inheritance of the resistance identified in the wild tomato species. For this purpose, resistant plants were selected and crossed to a susceptible cultivar, S. lycopersicum cv. Moneymaker to produce populations (usually F2 populations) for inheritance study. Crosses between S. lycopersicum and wild tomato species can be easy but sometimes require strategies such as embryo rescue, especially for the self‐incompatible species like S. peruvianum .
By using F2 populations, inheritance of resistance identified in several wild species was characterized. Monogenic resistance to O. neolycopersici was found in S. peruvianum LA2172, S. habrochaites G1.1560 and G1.1290, and polygenic resistance in S. neorikii G.16101. Rick, C.M. (1988). Further, by screening these F2 plants with molecular markers, such as RAPD, AFLP, and CAPS, the resistance in these species was mapped onto specific chromosomes. The resistance loci in S. peruvianum LA2172 and S. habrochaites G1.1560 and G1.1290, named Ol‐4 , Ol‐1, and Ol‐3 , respectively, are all located on tomato chromosome 6. The Ol‐4 locus is on the short arm, while Ol‐1 and Ol‐3 are on the long arm and closely linked if not allelic ( Figure B5.2). In addition to these monogenic Ol ‐genes, three quantitative trait loci (QTLs) were identified governing the resistance in S. neorickii G1.1601. The Ol‐qtl1 interval overlaps with Ol‐1 and Ol‐3 , while the other two linked Ol‐qtls are located on chromosome 12 in the vicinity of the Lv locus that confers resistance to another powdery mildew species, Leveillula taurica . Markers with close linkage to these loci were generated and can be applied in marker‐assisted selection (MAS) in breeding programs.
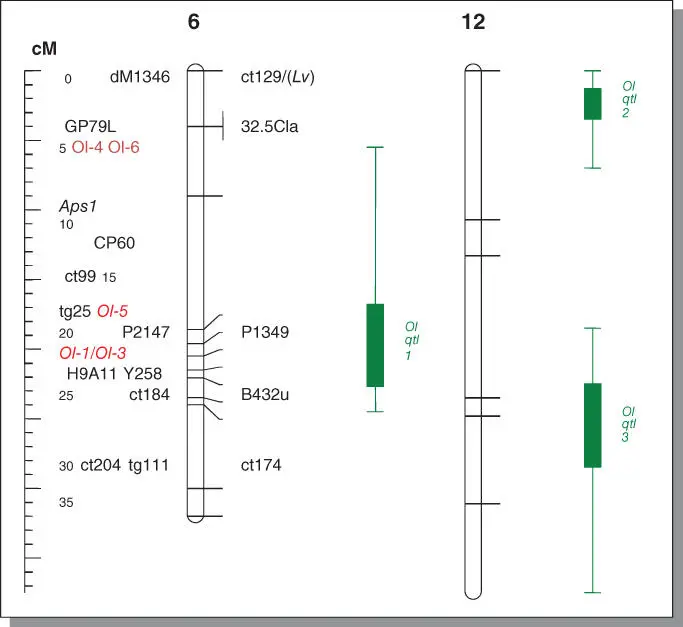
Figure B5.2The chromosome locations of tomato loci for resistance to tomato powdery mildew caused by Oidium neolycopersici . On the left, genetic distance in cM is shown. On the right, map positions of markers and resistance loci are shown on tomato chromosome 6 and 12, respectively. The donors for Ol‐1, Ol‐3, Ol‐5 are Solanum habrochaites G1. 1560, G1. 1290, and PI247087, respectively; for Ol‐4 is S. peruvianum LA2172 and for Ol‐qtls is S. neorickii G1.1601. Ol‐6 is identified from an advanced breeding line with unknown source. As to Ol‐qtls , bars indicate the QTL interval for which the inner bar shows a one‐LOD support and the outer one shows a two‐LOD support interval.
Generation of near isogenic lines
Near‐isogenic lines (NILs) that carry small introgressed chromosome fragments from related wild species in a cultivated tomato background are most useful pre‐bred in a breeding program. To develop NILs that only differ in Ol genes for resistance to O. neolycopersici, resistant donor accessions were crossed with susceptible S. lycopersicum cv. Moneymaker (MM). BC crosses were made starting from crossing F1 plants back to MM ( Figure B5.3). During the backcrossing, selection of resistant BC plants can be performed in two ways. One is by testing BC plants with powdery mildew and the other is to genotype them with markers linked to individual resistant loci. Since we have markers linked to each Ol‐ genes, we could apply MAS. We did a disease test since (i) it is a relatively easy disease assay which can be carried out at seedling stage; and (ii) the resistance phenotype is clear to be scored. In case that the disease assay is not easy to perform, for example due to (i) quarantine pathogens or (ii) a disease test that has be done at the late developmental stage, MAS would be a convenient way to select resistant plants ( Figure B5.4).
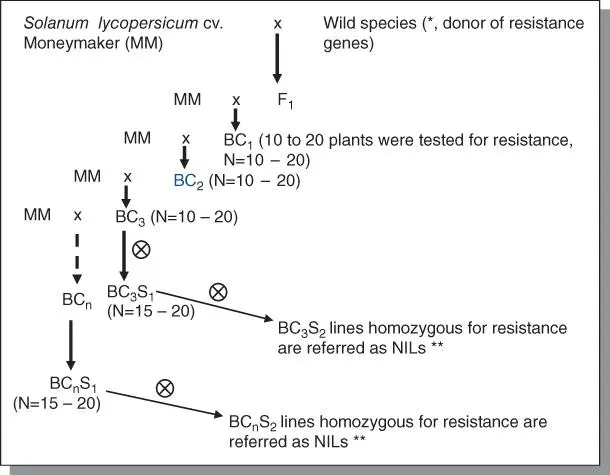
Figure B5.3Cross‐pollinating scheme of generation of near‐isogenic lines (NIL) harboring dominant resistance genes to tomato powdery mildew. Via backcrosses, new traits are introduced from wild tomato relatives. During the backcrosses, selection of resistant plants can be performed via (1) disease test and/or (2) marker‐assisted selection (MAS, Figure B5.4). In addition, select plants should have similar morphology to the recurrent parent. * and **: Usually it takes many generations to remove the deleterious genes that go along with the introduced genes due to linkage drag. Therefore, it is useful to start with advance breeding lines having introgression from the wild species in order to shorten the backcrossing procedure. In our practice, we used advance breeding lines derived from S. habrochaites G1.1560 (donor of the Ol‐1 gene) to produce the F 1. We checked BC 3S 1plants for the uniformity of their genetic background by genotyping these plants with 12 AFLP primer combinations that produce genome‐wide markers. Of the 30 AFLP marker alleles specific for S. habrochaites G1.1560, they were only present in the BC 3S 1plants with two fully cosegregating with the Ol‐1 gene, suggesting that the genetic background of these BC 3S 1plants are genetically similar to MM. For the Ol‐4 genes derived from S. peruvianum LA2172, we started with the wild accession. With 12 AFLP primer combinations, 48 AFLP marker alleles were identified from S. peruvianum LA2172 and 11 of these alleles still segregated in the tested BC 3S 1plants. Thus, when the backcross is started from wild accessions, more backcross generations are needed to make NILs.
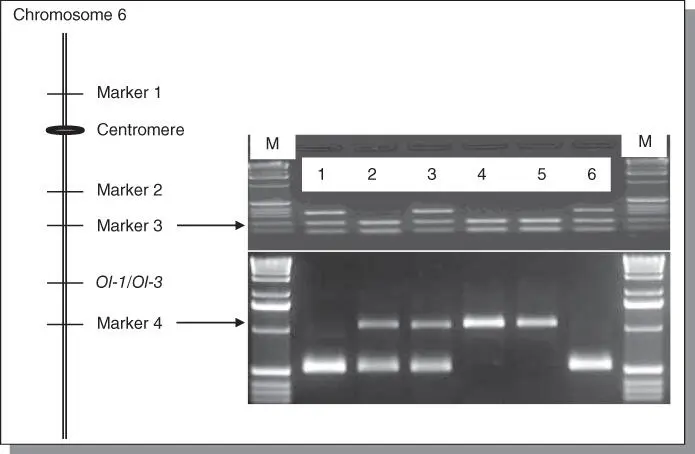
Figure B5.4Illustration of marker‐assisted selection (MAS). On the left, a genetic linkage map of tomato chromosome 6 showing that the Ol‐1 and Ol‐3 genes, conferring resistance to tomato powdery mildew, are located at the same locus and are flanked by Markers 3 and 4. On the right, electrophoretic patterns of PCR markers showing marker genotypes of 6 plants; the upper panel for Marker 3 and the lower panel for Marker 4. Plant 1–4 are either BC3 plants (for Marker 3) and BC3S1 plants (for Marker 4). Plant 5 and 6 are parental plants that are susceptible and resistant to tomato powdery mildew, respectively. M indicates DNA size marker of 1 kb ladder. For MAS, marker flanking the target gene is often used. For Marker 3, BC3 plants no. 1 and 3 are selected and expected to be resistant to powdery mildew since they have the marker allele of the resistant parent (plant no. 6). For Marker 4, plant 1 to 3 are selected and expected to be resistant since they have the resistant marker allele as the resistant parental plant no. 6 (homozygous plant no. 1 and heterozygous plant no. 2 and 3).
Читать дальше
















