aHaploid genome.
bIncluding protein‐coding and noncoding RNA genes.
Source: www.ebi.ac.uk/genomes.
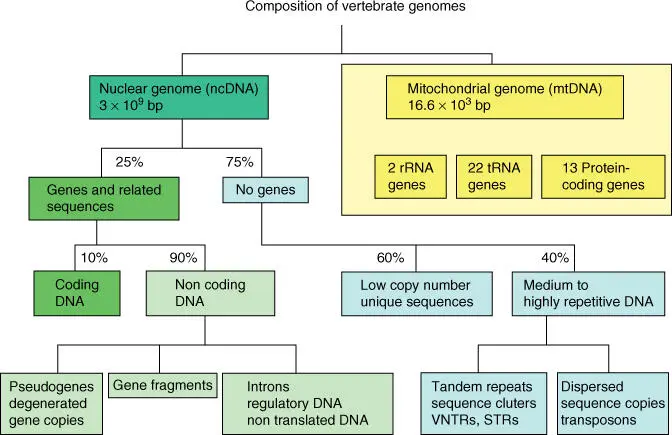
Figure 4.2 Composition of eukaryotic genomes and a fraction of a few DNA elements of the entire human genome.
Possibly the largest part of the genome (over 50% with higher eukaryotes) is not transcribed and according to our present knowledge is partially functionless. Important elements are pseudogenesand repetitive DNAsequences (Table 4.3and Figure 4.2).
Table 4.3 A few characteristics of the human genome.
| Parameter |
Human genome |
| Genome size |
3.2 × 10 9bp |
| Number of protein‐coding genes |
21 000 |
| Longest gene |
2.4 × 10 6bp |
| Medium gene size |
27 000 bp |
| Smallest number of exons/gene |
1 |
| Highest number of exons/gene |
178 |
| Mean number of exons/gene |
10.4 |
| Largest exon |
17 106 bp |
| Mean exon size |
145 bp |
| Number of pseudogenes |
>20 000 |
| Number of noncoding RNA genes |
>9000 |
| Percentage of protein‐coding sequences |
1.5% |
| Percentage DNA in rRNA, functional DNA |
3.5% |
| Percentage in repetitive DNA elements |
∼50% |
It is usually (but not always) the case that new functional genes develop through the doubling or duplication of genes in the progress of evolution. New genes can also be generated by combining domainsor partial gene sequences. Horizontal gene transfer(which happened when bacteria became mitochondria) also helped to enlarge the eukaryotic genomes. In contrast, pseudogenes, which are nontranslatable copies of genes, show frameshifts, nonsensemutations, deletions, and insertions (see Section 4.1.4). Pseudogenes do not have any further function today. Pseudogenescan be divided into two groups: the first arose from gene duplicationand the second from retroposons. In the second case, the genes were transcribed and processed and, following reverse translation in DNA, were inserted into a location in the genome. It is usually the case that these retropseudogeneshave no introns, but frequently poly(A) tails, and unlike the pseudogenes they are not present in the vicinity of the original gene from which they arose. Surprisingly, nature can afford to reproduce this junk DNA in every generation, even though replication is an energy‐consuming process. Perhaps these DNA sections that today appear to be useless will become functional in a later evolutionary phase as molecular replacement parts.
When the duplicated DNA sequence lies beside the original gene, it is termed as tandem repeat. These tandem repeats are the starting point for further DNA amplifications, induced by uneven crossing‐over. Repetitive DNAis quantitatively important and can be divided into middle repetitive DNA(transposons and retroelements) and highly repetitive DNA. The latter class includes short nucleotide sequences, which are present in great numbers in chromosomes in a tandem‐type style. There are also further divisions into telomere, satellite, minisatellite, and microsatellite DNA.
Upon cesium chloride gradient centrifugation, the DNA of eukaryotes is separated, and two bands are often observed, the smaller of which contains satellite DNA. This satellite DNAis especially rich in repetitive sequences and prefers to be localized in the region of the centromeres. In insects and other arthropods, this satellite DNA is very homogeneous, meaning that their sequence elements are highly conserved. In vertebrates the repeated sequence units contain up to 1000 repetitions of satellite DNA, and it is significantly longer and more variable (length of over 200 bp); subelements such as GA 5TGA can often be found in these elements. Through uneven crossing‐over, the variability of satellite DNA is about 10 times higher than with genes that only have a low copy number. Division and organization of the repetitive DNA elements in the centromere region are chromosome and type specific. It is assumed that the repetitive DNA at the centromere region is responsible for homologous chromosome recognition and the fact that they arrange themselves next to each other during meiosis.
In the actual satellite DNA of both plants and animals, elements are found that are repeated 5–50 times, each being 15–100 bp. The sequence elements can be attributed to the original sequence that was varied through point mutations. This repetitive DNA, each about 500–5000 nucleotides in length, is significantly shorter than the satellite DNA and is termed minisatelliteor variable number tandem repeat s ( VNTR s ). It exhibits a large variability in length in every locus, and a very high mutation rate is present as a result of uneven crossing‐over (as the number and length of repeats is changed), which can amount to 5% of the gamete. Minisatellite DNAis therefore termed the hot spotof meiotic recombination. Minisatellite DNA is especially suitable for the identification of individuals and has been used also for clarification of paternity and homozygosity in a population. Many VNTR loci each have dozens of alleles, which are codominantly inherited. This characteristic was used in DNA fingerprinting. The possibility that two unrelated individuals have the same DNA fingerprints is less than 1 in 10 million. Presently, DNA fingerprinting is based on short tandem repeat (STR) and single nucleotide polymorphism (SNP) analyses.
In addition, there are still shorter repeats that arise in animal and plant genomes. These consist of a basic unit of two (sometimes as many as five) nucleotides, such as (GC) nor (CA) n, which are repeated up to 100 times. Of these elements, termed microsatellitesor STRs (short tandem repeats ), about 30 000 loci are found in humans, which are of great importance for the recognition of tissues and individuals, paternity and population studies, and genome mapping. STR analysis is the method of choice for the determination of sexual crimes or murder in forensic medicine or criminal studies. The alleles allow amplification through polymerase chain reaction ( PCR )(see Chapter 13). Microsatellite PCRis currently the method of choice for many forensic, biotechnological, and biological investigations due to the fact that it requires only the smallest amounts of DNA. The variability of microsatellite DNA is strongly increased during meiosis via uneven crossing‐over and slippage of the DNA polymerase, so that the short sequence elements can be mutated, duplicated, and deleted. Alternatively to STR analyses, SNP analyseshave become available for a number of organisms, which often provide a more detailed picture of the genetic background.
Additional 500‐base long DNA sections are found in animal and plant genomes. These so‐called scattered or short interspersed element s ( SINE s), or 1000‐ to 5000‐nucleotide long interspersed element s ( LINE s), appear in high copy numbers (although not in tandem style repeats) ( Figure 4.2). The DNA elements Alu(which is recognized by the restriction enzyme Alu I), Kpn, and poly(CA)are also counted among the SINEs. The percentage of these elements in the human genome is about 20% of the entire genome. It is presumed that these elements, which are also called mobile genetic elementsor retrotransposons, arise through reverse transcription. From an evolutionary point of view, transposons (with long terminal repeat s [ LTR s]or inverted repeat s [ IR s]), retrotransposons, and retroposons (transposons without LTRs) could be considered as examples of active egoisticgenes ( selfish DNA), which only have their own replication in mind. On the other hand, these mobile elements lead to genetic variability (an increased exon shuffling or enhancer shuffling) that in the long run can also have positive effects. In areas of Alu sequences, chromosomes exhibit increased rates of new orientation. When Alu elements jump into active genes, most of them are inactivated; conversely, sleeping genes can be activated, in that the skipped elements can function as enhancers. Finally, the selection of new characteristics is made available. Sexual isolation and type formation can be increased through this mechanism.
Читать дальше




![Andrew Radford - Linguistics An Introduction [Second Edition]](/books/397851/andrew-radford-linguistics-an-introduction-second-thumb.webp)








