3 Krebs, J., Goldstein, E.S., and Kilpatrick, S.T. (2018). Lewin's Genes XII. Burlington: Jones & Bartlett Learning.
4 Biosynthesis and Function of Macromolecules (DNA, RNA, and Proteins)
Michael Wink
Heidelberg University, Institute of Pharmacy and Molecular Biotechnology (IPMB), Im Neuenheimer Feld 329, 69120, Heidelberg, Germany
4.1 Genomes, Chromosomes, and Replication
In the past few decades, genomicshas developed into a new specialized area of genetics and biotechnology. The aim is the complete molecular and functional characterization of genomes of all important organisms. It is divided into structural and functional genomics(see Chapter 21). When the human genome project HUGO (Human Genome Organization) determined the nucleotide sequence of a human haploid chromosome in 2001, this was a real breakthrough. Since then, sequencing technologies have changed. Instead of cloning and sequencing individual genes, today complete genomes are determined by massive parallel sequencing using next generation sequencing (NGS) (see Chapters 14and 21). More than 1150 other genomes are already completely sequenced (as of 2010), including 100 genomes of Eukarya, 970 of Bacteria, and 70 of Archaea (Table 4.1). In 2020, the number of species with sequenced genomes is higher than 10000 (see https://en.wikipedia.org/wiki/List_of_sequenced_bacterial_genomes, https://en.wikipedia.org/wiki/List_of_sequenced_eukaryotic_genomesfor an update) and new sequenced are published every eeek. By comparing nucleotide sequences and derived amino acid sequences obtained from various organ‐ and tissue‐specific cDNA and expressed sequence tag ( EST )banks, or through the construction of knockout RNAi, or antisense mutants, assigning the genomic sequences to functional units or genes is being attempted. Finally, functional genomics(see Chapters 21and 22) will supply an exact answer to the question of which regions of the genome have a function (today it is estimated that the information necessary for survival constitutes 85–95% of bacteria and only 10% of the whole DNA for vertebrates) and which parts can be regarded as apparently functionless evolutionary remnants. However, parts of the genome, which were considered functionless a few years ago, do have functions.
Table 4.1 Overview of a few of the genomes that are already sequenced and published.
| Organism |
Size (Mb) |
| Archaebacteria |
| Archaeoglobus fulgidus |
2.18 |
| Methanobacterium thermoautotrophicum |
1.75 |
| Methanococcus jannaschii |
1.66 |
| Pyrococcus horikoshii |
1.80 |
| Eubacteria |
| Bacillus subtilis (Gram‐positive bacterium) |
4.21 |
| Borrelia burgdorferi (borreliosis pathogen) |
1.44 |
| Chlamydia trachomatis (pathogen of urogenital tract) |
1.05 |
| Escherichia coli (intestinal bacterium) |
4.64 |
| Haemophilus influenzae (pathogen of purulent throat infections) |
1.83 |
| Helicobacter pylori (stomach ulcer pathogen) |
1.67 |
| Mycobacterium tuberculosis (tuberculosis pathogen) |
4.45 |
| Mycoplasma pneumoniae (pneumonia pathogen) |
0.81 |
| Rickettsia prowazekii (typhus fever pathogen) |
1.10 |
| Treponema pallidum (syphilis pathogen) |
1.14 |
| Eukaryotes |
| Plasmodium falciparum (malaria pathogen) |
1.00 |
| Saccharomyces cerevisiae (Brewer's yeast) |
12.069 |
| Arabidopsis thaliana (Arabidopsis) |
220 |
| Caenorhabditis elegans (nematode) |
130 |
| Drosophila melanogaster (fruit fly) |
200 |
| Mus musculus (house mouse) |
2800 |
| Homo sapiens (human) |
3200 |
Mb, one million bases.
The total DNA of a cell is referred to as a genome. Genome sizes of major organismal groups are shown schematically in Figure 4.1. When the minimal genome size of organisms is examined (i.e. only the left side of the bar), an increase in size can be seen that mainly runs parallel to the organizational level. Bacteria and fungi with simple structures have smaller genomes than structurally complicated multicellular organisms. It is presumed that the genome was enlarged particularly through genome duplications. Protostomia and the Deuterostomia ancestors of the vertebrates (see Chapter 6) contain generally only one copy of a gene, while several copies of a gene are often found in the genomes of chordates. As a result, it is supposed that the chordate genomes have doubled at least two or three times ( 1‐2‐4 rule). The first genome duplication during the evolution of chordateshas already taken place before the Cambrian explosion, whereas the second and next doubling occurred in the early Devonian period. In the evolution of fish, a further doubling of the genome occurred with up to eight copies of the original Deuterostomia (1‐2‐4‐8 hypothesis) in the late Devonian period. This took place after the Actinopterygii and Sarcopterygii had already divided. Among the Sarcopterygii are the famous Coelacanthus and lungfishes. All land vertebrates (amphibians, reptiles, birds, and mammals) have apparently descended from them. Within the eukaryotes, the maximum genome size has only a small relationship to the developmental level. This is because many plants and amphibians have genomes with up to 10 11bases, and the genomes are therefore one to two orders of magnitude higher than the genome of humans – it is obvious that many genome duplications must have taken place in these groups.
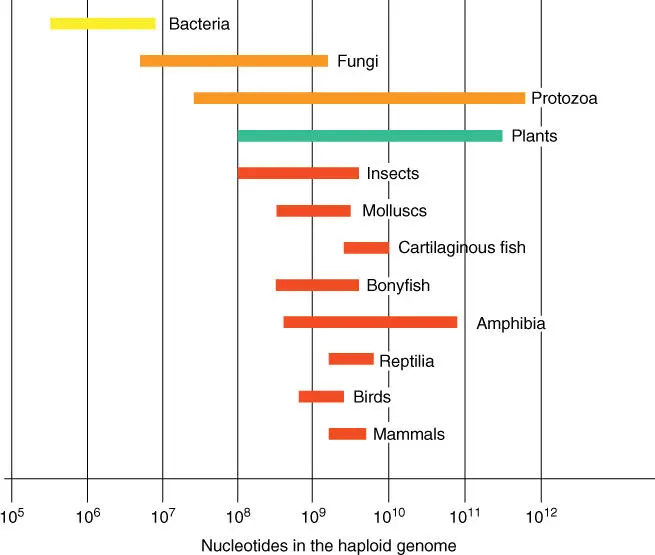
Figure 4.1 Number of nucleotides in the haploid genomes of important groups of organisms.
When the human genome is considered, it is obvious that a massive amount of information is present. If the DNA in an individual human cell was stretched out, it would be 2 m long. With around 10 13cells in our body, the total length of DNA in all cells is 2 × 10 10km. This length would be a distance that runs many times from the earth to the sun and back again!
Of the 3.2 million bases that are present in human haploid chromosomes, about 25% of the DNA defines genes, but only 1.5% of the DNA codes directly for proteins (Table 4.2and Figure 4.2). The rest of the DNA is made up of RNA genes and noncoding sequences, which often either serve no function or their function is still unknown. In recent years microRNAs have been detected encoded in the “functionless” DNA, which are important for gene regulation (see Chapters 3and 21).
Table 4.2 Relation between genome size and the number of genes of a few selected species whose genomes have been sequenced.
| Organisms |
Genome size (bp) a) |
Approximate number of genes b) |
| Archaea |
| Archaeoglobus fulgidus |
2.18 × 10 6 |
2405 |
| Methanothermobacter thermautotrophicus |
1.75 × 10 6 |
1866 |
| Pyrococcus furiosus (Archaea) |
1.91 × 10 6 |
2057 |
| Sulfolobus acidocaldarius (Archaea) |
2.99 × 10 6 |
2221 |
| Bacteria |
| Clostridium tetani |
2.8 × 10 6 |
2373 |
| Escherichia coli |
4.67 × 10 6 |
4288 |
| Haemophilus influenzae |
1.83 × 10 6 |
1702 |
| Mycoplasma genitalium |
0.58 × 10 6 |
476 |
| Rhodospirillum rubrum |
4.35 × 10 6 |
3791 |
| Fungi |
| Aspergillus fumigatus |
2.9 × 10 7 |
9920 |
| Saccharomyces cerevisiae |
1.3 × 10 7 |
6600 |
| Candida glabrata |
1.4 × 10 7 |
5180 |
| Sporozoa |
| Plasmodium falciparum (causes malaria) |
2.3 × 10 7 |
5300 |
| Plants |
| Arabidopsis thaliana |
2.2 × 10 8 |
29000 |
| Animals |
| Caenorhabditis elegans (nematode) |
1.3 × 10 8 |
21 000 |
| Drosophila melanogaster (fruit fly) |
2.0 × 10 8 |
32 000 |
| Danio rerio (zebra fish) |
1.4 × 10 9 |
21 000 |
| Mus musculus (mouse) |
2.8 × 10 9 |
30 000 |
| Homo sapiens (human) |
3.2 × 10 9 |
30 000 |
((done))
Читать дальше




![Andrew Radford - Linguistics An Introduction [Second Edition]](/books/397851/andrew-radford-linguistics-an-introduction-second-thumb.webp)








