Source: Based on Stojanovic et al. [14].
The RCDZ logic gates can be connected via cascades of deoxyribozyme‐catalyzed reactions. For example, Stojanovic et al. connected YES, NOT, AND, and ANDNOT logic gates to downstream YES RCDZ gate [40]. An example of 2iAND gate is shown in Figure 4.2. Deoxyribozyme ligase‐based 2iAND gate, when activated by the two DNA inputs I Aand I B, can bind the two short strands OUTa and OUTb and covalently link them into a longer oligonucleotide OUT ( Figure 4.2c). The latter can be recognized by a downstream RCDZ gate, as shown in Figure 4.2d. Such cascade resulted in a two‐layer logic gate integration. Incubation period for up to 60 minutes was required for this system to achieve fluorescent response above the background. The disadvantage of the system is slow release of the output oligonucleotide from the complex with the ligase gate, since the product of ligation has higher affinity to the DNA ligase than the substrates (OUTa and OUTb).
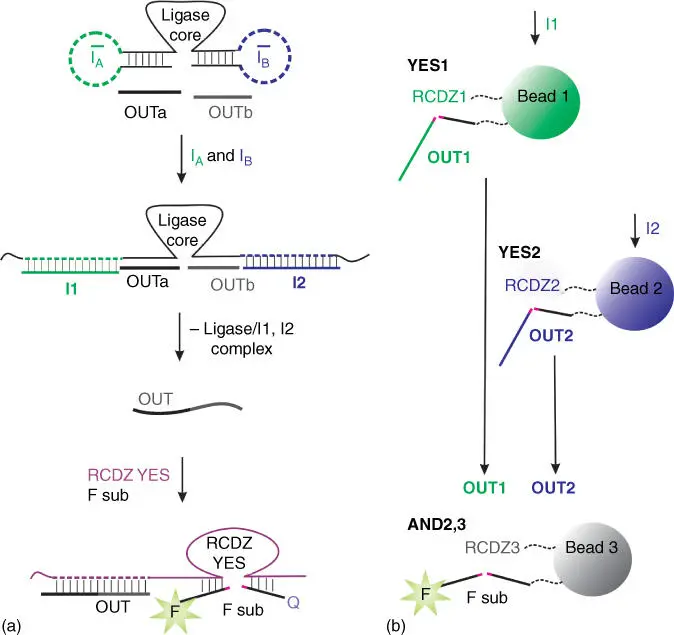
Figure 4.2AND deoxyribozyme ligase gate connected to YES RCDZ gates. (a) DNA ligase‐based two‐input AND gate: the ligase Dz is inhibited by two stem‐loop structures. Inputs I Aand I Bunwind the inactivating stem‐loops and enable the DNA ligase to ligate strands OUTa and OUTb to produce the OUT strand. The OUT strand activates RCDZ for cleavage of F sub followed by fluorescence increase. (b) Beads 1 and 2 contain immobilized RCDZ constructs and their corresponding substrates. They act as YES logic gates: in the presence of the oligonucleotide inputs I1 and I2, they activate RCDZ and release OUT1 and OUT2 oligonucleotides. The released OUT1 and OUT2 can activate AND2,3 gate on the third bead, which can be monitored via fluorescence.
Yashin et al. immobilized RCDZ constructs on microsphere beads together with their substrates ( Figure 4.2b) [41]. The substrates were inactivated by complementary strands, which could be removed by inputs I1 and I2, respectively (not shown in Figure 4.2b). The YES beads, activated by certain input combination, could release their oligonucleotide outputs in solution (OUT1 and OUT2), which then could be recognized by downstream logic gates (AND2,3 gate in Figure 4.2b). Three-layer integration was achieved. The advantage of the approach is in the ability to monitor the fluorescent signal from individual beads by flow cytometry. However, hybridization of the bead‐immobilized oligonucleotides might be significantly slower than that in solution [42]. Indeed, time needed to observe a strong signal on the last bead in this case was 16 hours [41].
Bone et al. used split cascades based on the most catalytically active 10–23 Dz that enable realizing inactivated RCDZ [43]. This approach can reduce the amount of input required for cascade activation from 20–1000 nM to 20–100 pM [43,37,44].
4.3 Connecting Gates Based on DNA Strand Displacement
The design of seesaw gates [53] takes advantage of DNA strand displacement, e.g. the ability of a partial DNA duplex to release one strand upon hybridization of the second strand with an interfering strand that can form more Watson–Crick base pairs than presented in the original duplex ( Figure 4.3a). The phenomenon has been used as a probe for the detection of specific nucleic acids in several variations [45–52]. For example, a DNA duplex composed of a 5′‐fluorophore‐labeled and a 3′‐quencher‐labeled oligonucleotide strands can be interrogated by a complementary analyte that “pushes” the fluorophore‐containing strand in solution ( Figure 4.3a), which can be reported as a fluorescent signal in a quantitative real‐time polymerase chain reaction (PCR) format [48].
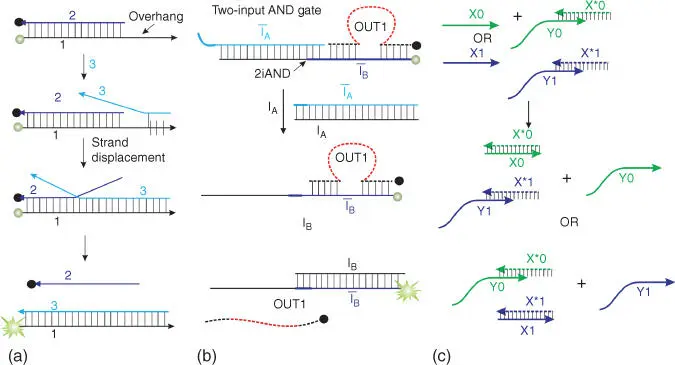
Figure 4.3Design of strand displacement (seesaw) DNA logic gates [53]. (a) Strand displacement‐based sensor for nucleic acid detection. (b) Two‐input AND logic gate, which consist of the complex ī Aand ī Bwith OUT1, releases OUT1 as an output upon binding to inputs ī Aand ī B. Red dashed line indicates unique sequence independent on the sequence of ī Aand ī Bstrands. (c) Dual‐rail logic. YES gate made with rail logic. X0 and X1 represent negative or positive input, respectively. Y0 and Y1 represent negative or positive output signal, respectively.
The advantages of this system for DNA logic gate design are the following:
1 (i) Design simplicity.
2 (ii) The double‐stranded constructs with only short single‐stranded overhangs ( toeholds) reduce the nonspecific associations between oligonucleotides, which enables usage of many different strands in the same solution.
3 (iii) Both input and output signals are specific DNA sequences, which enables building cascades of communicated logic gates.
Seelig et al. designed a series of logic gates including AND, NOT, OR, thresholder, and amplifier [53]. A general idea for the design of a 2iAND gate is illustrated by Figure 4.3b. Strand ī Bforms a complex with strands ī Aand OUT1. Strand OUT1 is released free in solution only in the presence of the two oligonucleotide inputs I Aand I B, which bind strands ī Aand ī B, respectively, forming the “waste” duplex products. Strand OUT1 can then serve as a unique input for the downstream gates due to the presence of a unique sequence (shown as a red fragment in Figure 4.3b). Using this strategy, Seelig et al. demonstrated a five‐layer DNA gate integration, which consisted of 11 gates and accepted six inputs [53]. A systematic construction of strand displacement gates can lead to up to 78 gates performing simultaneously [54]. The gates can be used for solving as complex tasks as mimicking natural neural networks [55]. The disadvantages of strand displacement gates include the following: (i) The intensity of signal is proportional to strand concentration, unlike that of RCDZ, in which signal is enhanced by the catalytic action of RCDZ. (ii) DNA interactions are thermodynamically driven, which leads to irreversibility of the circuits or continuous accumulation of the DNA waste products in each operational round. (iii) Strand displacement is slower than hybridization of two single‐stranded oligonucleotides.
Instead of interpreting presence of a particular strand as a positive signal and absence of the strand as a negative signal, one can use two sequences as positive and negative signals, respectively. Such method is called dual‐rail logic ( Figure 4.3c) [56]. Using this method, any gate that consists of AND/OR/NOT gates can be redesigned by using AND and NOT gates [56]. This approach was used for making logic gates suitable for both cascading and multiple operations [56,57]. Specific sequence complementary to one of the signal strands can be introduced in the solution, bind the input sequence, decompose the gate–input complex, and reactivate the gates. It leads to reassociation of the gate with its outputs. After that, a new input can be introduced into the computational system, with the ability to generate the proper output being saved.
The response rate of a DNA computational unit is limited by the time required for an input sequence and a gate to encounter each other in solution ( Figure 4.4). To mitigate this limitation, the logic gates can be localized at a short distance from each other, which allows for faster inter‐gate communication [27]. A scaffold/substrate, suitable for gate immobilization, can be a DNA tile [27]. Chatterjee et al. localized strand displacement DNA logic gates on a DNA origami tile ( Figure 4.4) [58]. Up to eight hairpins could communicate with each other [58], suggesting a possibility to integrate eight layers of DNA logic gates. Co‐localization of the circuit elements decreased the computation time from hours to minutes, as compared with solution‐based seesaw circuits.
Читать дальше














