Microsatellite analysis has also been used to identify the strains of S. uvarum (Masneuf‐Pomarède et al ., 2007, 2016) and of S. kudriavzevii (Erny et al ., 2012) used in winemaking. As the S. uvarum , S. kudriavzevii , and S. cerevisiae microsatellites have different amplification primers, this method provides an additional means of distinguishing between these species and their hybrids.
The development of new‐generation sequencing methods for yeast genomes has made sequences available for non‐Saccharomyces species. A typing method of yeast strains by microsatellite marker analysis is now offered for B. bruxellensis , T. delbrueckii , H. uvarum , and Starmerella bacillaris (Albertin et al ., 2014a,b, 2016; Masneuf‐Pomarede et al ., 2015, 2016). When applied to the study of a great number of yeast isolates, these methods help to better describe the genetic diversity and the population structure of winemaking yeasts. Factors influencing this structure as well as their life cycle and reproduction mode have also been described. From an applied point of view, this molecular typing method is a useful tool in winemaking yeast strain identification, ecological surveys, and quality control of industrial production batches.
With the development of new‐generation sequencing methods, new approaches to yeast characterization have been suggested. The multilocus sequence typing (MLST) method is a standardized approach to full or partial sequence analysis of certain gene expressions in yeast. These genes are characterized by a slow accumulation of mutations, which help differentiate between individuals, as well as deduce phylogenetic relationships between strains. Applied to S. cerevisiae , the results obtained do not indicate a superior ability to discriminate among yeast strains when compared with analysis by repetitive‐element PCR or microsatellite marker polymorphism (Ayoub et al ., 2006). However, studies have revealed the specific population structure of wine yeasts, confirming the domestication of these yeasts (Fay and Benavides, 2005). Other approaches consist in establishing sequences of regions located between randomly selected restriction sites in the genome (restriction site‐associated DNA sequencing or RAD‐seq). Many positions of variation of a base, or single nucleotide polymorphism (SNP), can thus be used for phylogenetic analyses. The RAD‐seq method has established the diversity and genetic structure of S. cerevisiae strains from a variety of ecological niches (Cromie et al ., 2013; Hyma and Fay, 2013).
1.10 Ecology of Grape and Wine Yeasts
1.10.1 Succession of Grape and Wine Yeast Species
A large amount of research was focused on the description and ecology of wine yeasts. It concerned the distribution and succession of species found on the grape and then in wine during fermentation and conservation (Ribéreau‐Gayon et al ., 1975; Lafon‐Lafourcade, 1983).
The ecological study of grape and wine yeast species represents a considerable amount of research. De Rossi began his research in the 1930s (De Rossi, 1935). Castelli (1955, 1967) pursued the study of yeast ecology in Italian vineyards. Peynaud and Domercq (1953) and Domercq (1956) published the first results on the ecology of wine yeasts in France. They described not only the species found on the grape and during alcoholic fermentation but also contaminating and spoilage yeasts. Among the many publications on this topic since the 1960s in viticultural regions around the world, the following works are worth noting: Brechot et al . (1962), Sapis‐Domercq (1970), Barnett et al . (1972), Minarik (1971), Cuinier and Guerineau (1976), Park (1975), Soufleros (1978), Belin (1979, 1981), Poulard et al . (1980), Poulard and Lecocq (1981), Bureau et al . (1982), Rossini et al . (1982), Fleet et al . (1984), Mills et al . (2002), Baleiras Couto et al . (2005), Hierro et al . (2006), and Nisiotou and Nychas (2007).
Yeasts are widespread in nature and are found in soils, on the surface of plants, and in the digestive tract of animals. Wind, insects, and birds disseminate them. Yeasts are distributed irregularly on the surface of the grapevine; found in small quantities on leaves, the stems, and unripe grapes, they colonize the grape skin during maturation. During the winter season, the main natural reservoir of yeasts is the trunk, while during the vegetative growth phase of the vine, it is found in the ground and in berries (Cordero‐Bueso et al ., 2011). Observations under a scanning electron microscope have identified the location of yeasts on the grape. They are rarely found on the bloom, but multiply preferentially on exudates released from microlesions in zones situated around the stomatal apparatus. Botrytis cinerea spores and lactic acid and acetic acid bacteria also develop in the proximity of these peristomatic fractures ( Figure 1.34).
The number of yeasts on the grape berry, just before harvest, is between 10 3and 10 5, depending on the geographical location of the vineyard, weather conditions during maturation, the healthiness of the harvested grapes, and pesticide treatments applied to the vine. The most abundant yeast populations are obtained under warm weather conditions (lower latitudes and higher temperatures). Insecticide treatments and certain fungicidal treatments can make the native grape microflora less populous. Quantitative results available on this subject, however, are few. After the harvest, transport, and crushing of the crop, the number of cells capable of forming colonies on an agar medium generally reaches 10 6cells/ml of must.
The number of yeast species significantly present on the grape is limited. From fruit set to maturity, Aureobasidium pullulans , Cryotococcus , and Rhodotorula are the main genera and species encountered (Prakitchaiwattana et al ., 2004; Martins et al ., 2014). Their proportion then decreases in favor of Ascomycetes. Among the latter, the apiculate species ( K. apiculata and its sporogenous form H. uvarum ) are the most common. They comprise up to 99% of the yeasts isolated in certain grape samples. The following are generally found but in lesser proportions: Metschnikowia pulcherrima , C. famata , C. stellata , P. membranifaciens , Pichia fermentans , and Hansenula anomala . The increase of grape exudate sugar content may partly explain these changes in yeast populations (Martins, 2012).
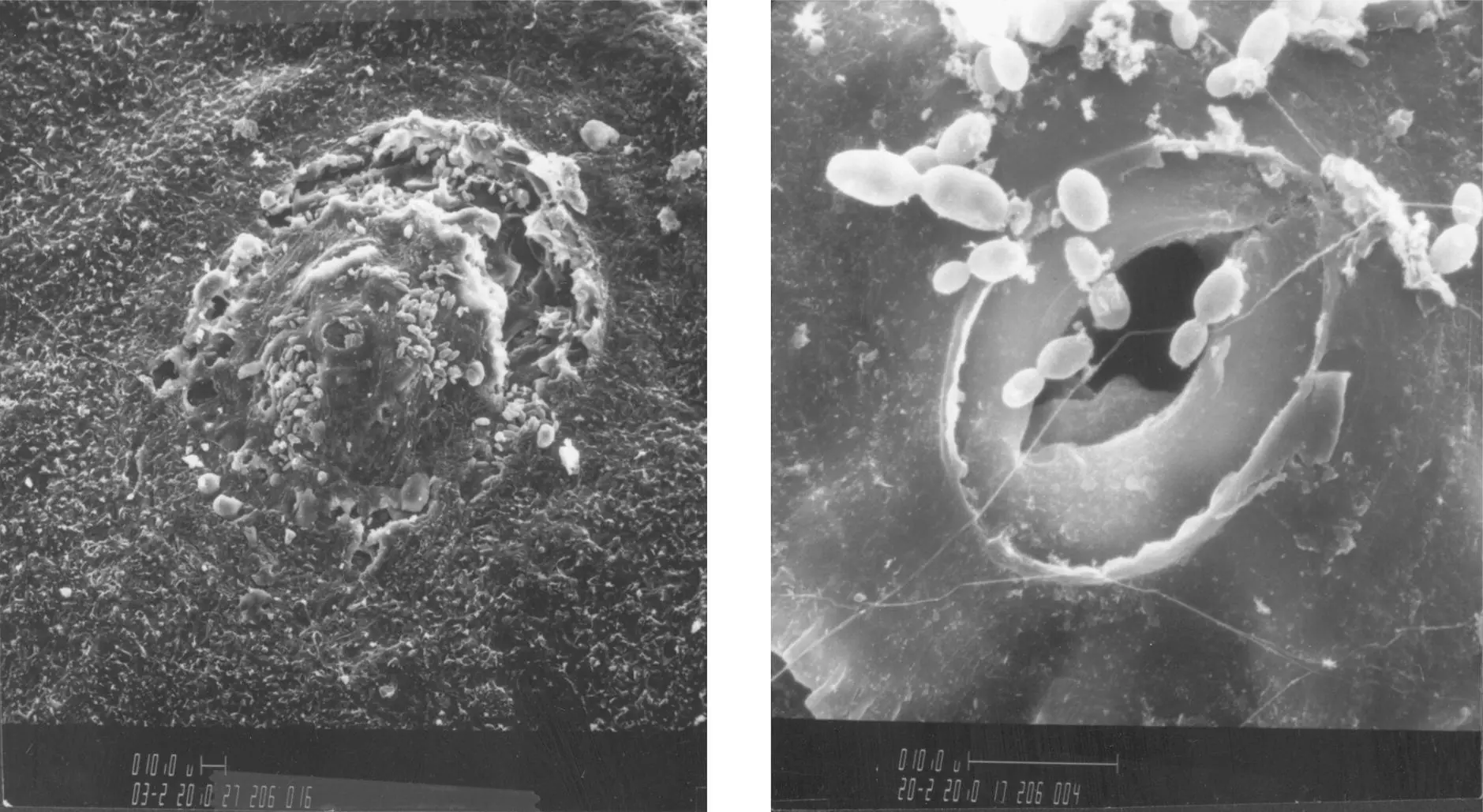
FIGURE 1.34 Grape surface under scanning electron microscope, with detail of yeast peristomatic zones.
( Source: Photographs from B. Pucheu‐Plante and M. Mercier, Department of Electron Microscopy, Université de Bordeaux 1.)
All research confirms the extreme rarity of S. cerevisiae on grapes (Mortimer and Polsinelli, 1999). However, these yeasts are not totally absent. Their existence cannot be proven by streaking diluted must on a solid medium prepared under aseptic conditions, but their presence on grapes can be proven by analyzing the spontaneous fermentative microflora of grape samples placed in sterile bags, then aseptically crushed and vinified in the laboratory in the absence of all contamination. Red and white grapes from the Bordeaux region were treated in this manner. At mid‐fermentation in the majority of cases, S. cerevisiae represented almost all of the yeasts isolated. In some rare cases, no yeast of this species developed and non‐ Saccharomyces yeasts began the fermentation. Nevertheless, growth of S. cerevisiae under these conditions from healthy grapes is not guaranteed. Thus, based on 134 samples of grapes collected in the Bordeaux region, 31% of samples were positive for S. cerevisiae at the two‐thirds mark of alcoholic fermentation (Börlin, 2015). Under the same operating conditions, 28–38% of positive fermentation was obtained for grape samples from the Douro region in Portugal (Schuller et al ., 2012). The frequency of isolation of S. cerevisiae may approach 100% on damaged grapes, which are a very favorable environment for the development of fermentation yeasts (Mortimer and Polsinelli, 1999).
Читать дальше













