The extraction of mtDNA comprises several stages. The protoplasts obtained by enzymatic digestion of the cell walls are lysed in a hypotonic buffer. The mtDNA is then separated from the chromosomal DNA by ultracentrifugation in a cesium chloride gradient, in the presence of bisbenzimide, which acts as a fluorescent intercalating stain. This agent amplifies the difference in density between chromosomal DNA and mtDNA. The mtDNA has a high number of adenine and thymine base pairs, for which bisbenzimide has a strong affinity. Finally, the mtDNA is purified by a phenol–chloroform extraction and an ethanol precipitation.
Defontaine et al . (1991) and Querol et al . (1992) simplified this protocol by separating the mitochondria from the other cell constituents before extracting the DNA. In this manner, they avoided the ultracentrifugation step. The coarse cellular debris is eliminated from the yeast lysate by centrifuging at 1,000 g. The supernatant is then recentrifuged at 15,000 g to obtain the mitochondria. The mitochondria are then lysed in a suitable buffer to liberate the DNA.
Unlike the industrial brewer's yeast strains analyzed by Aigle et al . (1984), which have the same mtDNA restriction profile, implying that they are of common origin, the winemaking yeast strains have a large degree of mtDNA diversity. This method easily differentiates between most of the selected yeasts used in winemaking as well as wild strains of S. cerevisiae found in spontaneous fermentations ( Figure 1.26). This method may also help differentiate S. uvarum strains (Naumova et al ., 2010).
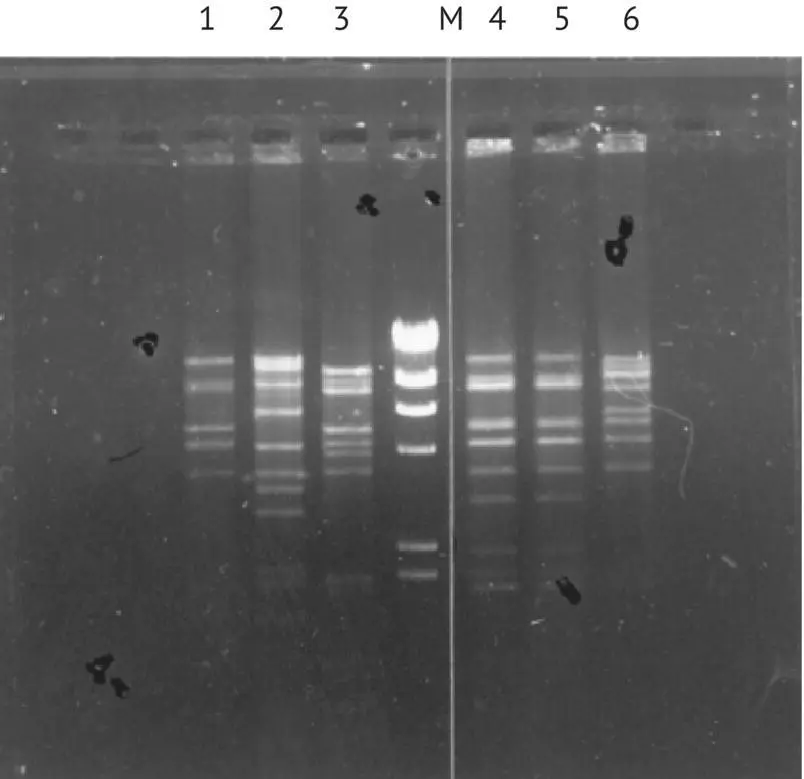
FIGURE 1.26 Restriction profile by Eco R5 of mtDNA of different strains of S. cerevisiae . Band 1, FIO; band 2, BO213; band 3, VLI; M, marker; band 4, 522; band 5, Sita 3; band 6, VL3c.
This technique is very discriminating and not too expensive, but it is long and requires several complex manipulations. It is useful for the subtle characterization of a small number of strains. Inoculation effectiveness can also be verified by this method. In the laboratory, the lees, sampled during or toward the end of alcoholic fermentation, are cultured in a liquid medium. The mtDNA restriction profiles of this total biomass and of the yeast starter strain are compared. The absence of any extra bands, with respect to the yeast starter strain restriction profile, demonstrates that the yeast starter has been properly implanted, with an accuracy of 90%. In fact, in the case of a binary mixture, the minority strain must represent around 10% of the total population to be detected (Hallet et al ., 1989).
Saccharomyces cerevisiae has 16 chromosomes with a size range between 250 and 2,500 kb. Its genomic DNA is very polymorphic; thus, it is possible to differentiate strains of the species according to the size distribution of their chromosomes. Pulsed‐field electrophoresis is used to separate S. cerevisiae chromosomes and to compare karyotypes of the strains. This technique uses two electric fields oriented differently (90–120°). The electrodes placed on the sides of the apparatus apply the fields alternately ( Figure 1.27).
FIGURE 1.27 CHEF pulsed‐field electrophoresis device ( contour clamped electrophoresis field ).
The user can define the duration of the electric current that will be applied in each direction (pulse). With each change in the direction of the electric field, the DNA molecules reorient themselves; the smaller chromosomes reorient themselves more quickly than the larger ones ( Figure 1.28).
Blondin and Vezhinet (1988), Petering et al . (1988), and Dubourdieu and Frezier (1990) applied this technique to identify wine yeast strains. Sample preparation is relatively easy. The yeasts are cultivated in a liquid medium, collected during the log phase, and then placed in suspension in a warm agarose solution that is poured into a partitioned mold to form small plugs.
Figure 1.29gives an example of the identification of S. cerevisiae strains isolated from a grape must undergoing spontaneous fermentation. Vezhinet et al . (1990) have shown that karyotype analysis can distinguish between strains of S. cerevisiae as well or better than the use of mtDNA restriction profiles. Furthermore, karyotype analysis is much quicker and easier to use than mtDNA analysis. In the case of ecological studies of spontaneous fermentation microflora, pulsed‐field electrophoresis of chromosomes is extensively used today to characterize strains of S. cerevisiae (Frezier and Dubourdieu, 1992; Versavaud et al ., 1993, 1995).
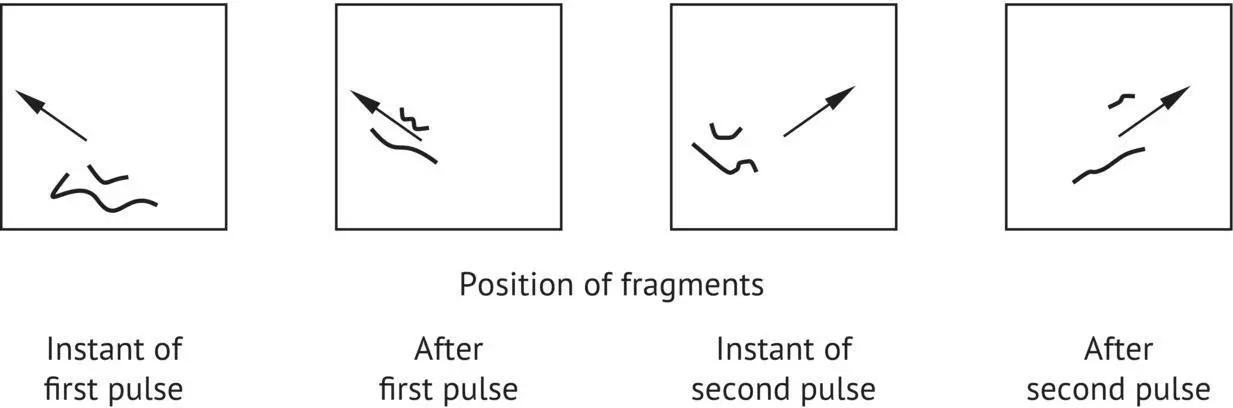
FIGURE 1.28 Mechanism of DNA molecule separation by pulsed‐field electrophoresis.
Very little research on the chromosomal polymorphism in other species of grape and wine yeasts is currently available. Naumov et al . (1993) suggested that S. uvarum and S. cerevisiae karyotypes can be easily distinguished. Other authors (Vaughan Martini and Martini, 1993; Masneuf, 1996) have confirmed their results. In fact, a specific chromosomal band systematically appears in S. uvarum . Furthermore, there are only two chromosomes whose sizes are less than 400 kb in S. uvarum but generally more in S. cerevisiae , in all of the strains that we have analyzed.
Non‐ Saccharomyces species, in particular apiculate yeasts ( H. uvarum and K. apiculata ), are present on grapes and are sometimes found at the beginning of fermentations. These species have fewer polymorphic karyotypes and fewer bands than in Saccharomyces . Versavaud et al . (1993) differentiated between strains of apiculate yeast species and Candida famata by using restriction endonucleases at rare sites ( Not 1 and Sfi 1). The endonucleases cut the chromosomes into a limited number of fragments, which were then separated by pulse‐field electrophoresis.
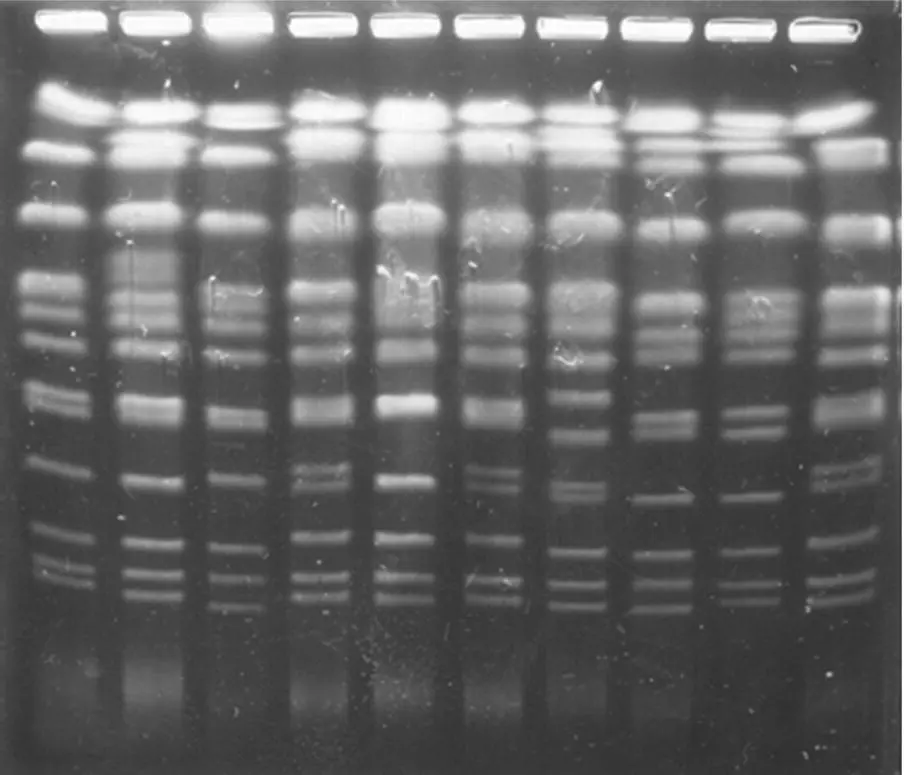
FIGURE 1.29 Example of electrophoretic (pulsed field) profile of S. cerevisiae strain karyotypes.
1.9.4 Genomic DNA Restriction Profile Analysis Associated with DNA Hybridization by Specific Probes (Fingerprinting)
The yeast genome contains DNA sequences that repeat from dozens to hundreds of times, such as the δ sequences or Y1 elements of the chromosome telomeres. The distribution, or more specifically, the number and location of these elements, has a certain intraspecific variability. This genetic fingerprint is used to identify strains (Pedersen, 1986; Degre et al ., 1989).
The yeast strains are cultivated in a liquid medium and are sampled during the log phase, as in the preceding techniques. The entire DNA is isolated and digested by restriction endonucleases. The generated fragments are separated by electrophoresis on agarose gel and then transferred to a nylon membrane (Southern, 1975). Complementary radioactive probes (nucleotide sequences taken from δ and Y1 elements) are used to hybridize with fragments having homologous sequences. The result gives a hybridization profile containing several bands.
Читать дальше
















