Cheanyeh Cheng - Enzyme-Based Organic Synthesis
Здесь есть возможность читать онлайн «Cheanyeh Cheng - Enzyme-Based Organic Synthesis» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Enzyme-Based Organic Synthesis
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:3 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 60
- 1
- 2
- 3
- 4
- 5
Enzyme-Based Organic Synthesis: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Enzyme-Based Organic Synthesis»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
An insightful exploration of an increasingly popular technique in organic chemistry Enzyme-Based Organic Synthesis
Enzyme-Based Organic Synthesis
Enzyme-Based Organic Synthesis
Enzyme-Based Organic Synthesis — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Enzyme-Based Organic Synthesis», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
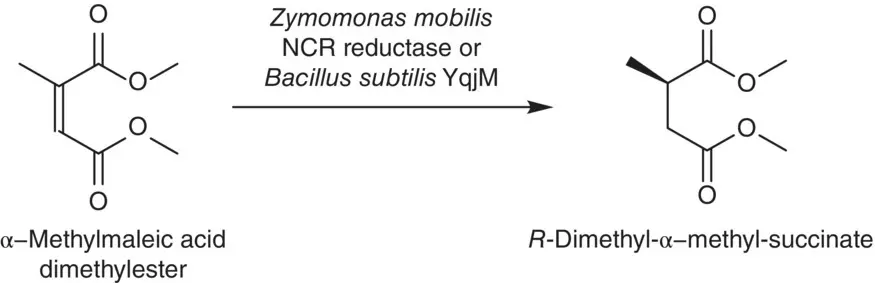
Scheme 2.41 Asymmetric reduction of α‐methylmaleic acid dimethylester with Z. mobilis or B. subtilis .
A novel ER isolated from the bacterium Z. mobilis termed NCR reductase and OYEs 1–3 from yeast of Saccharomyces spp. were extended to use for the asymmetric reduction of a variety of activated C=C substrates. The activating groups in the investigated substrates include aldehyde, ketone, imide, nitro, carboxylic acid, and ester moieties. To control the stereospecificity of the reaction, strategies such as variation of the main substrate structure (cyclic vs. acylic), switching the ( E / Z ) configuration of the alkene moiety, modifying the substitution pattern (mono‐ vs. di‐, α‐ vs. β‐), or proper selection of the enzyme are utilized to allow the access to the opposite enantiomeric products. Results showed that reaction rates and stereoselectivities can be varied by different substrate type. The “substrate‐based stereocontrol” is related with the ring size of cycloalkenones, position of substituents on the C=C bond, and its E / Z configuration. The “enzyme‐based stereocontrol” was observed with nitroalkene in which the opposite enantiomeric products can be obtained by both NCR reductase and OYEs 1–3 [184].
Baker’s yeast (BY) catalyzed enantioselective reduction of an EWG substituent activated double bond such as α,β‐unsaturated aldehydes and ketones are generally very efficient with high enantioselectivities and conversions. However, it is necessary to explore the synthetic capability of the yeast‐based ER for other activated substrates to make the enantioselective synthesis toolbox more fruitful and advantageous for implementation in industrial manufacturing processes. Therefore, the enantioselective reduction of the activated C=C substrates on some ( Z )‐methyl α‐halo‐β‐arylacrylates was investigated and performed by baker’s yeast fermentation or OYEs 1–3 mediated biotransformations to prepare ( S )‐α‐halo‐β‐arylpropionic acid derivatives in high e.e. and with good yields. The fermentation medium promotes ester hydrolysis, but the isolated enzymes preserve the ester functionality ( Scheme 2.43). Particularly, when aromatic ring was substituted by an EWG, high e.e. and conversion values were observed [185].

Scheme 2.42 Asymmetric bioreduction of citraconic acid dimethylester via a coupled‐substrate system.
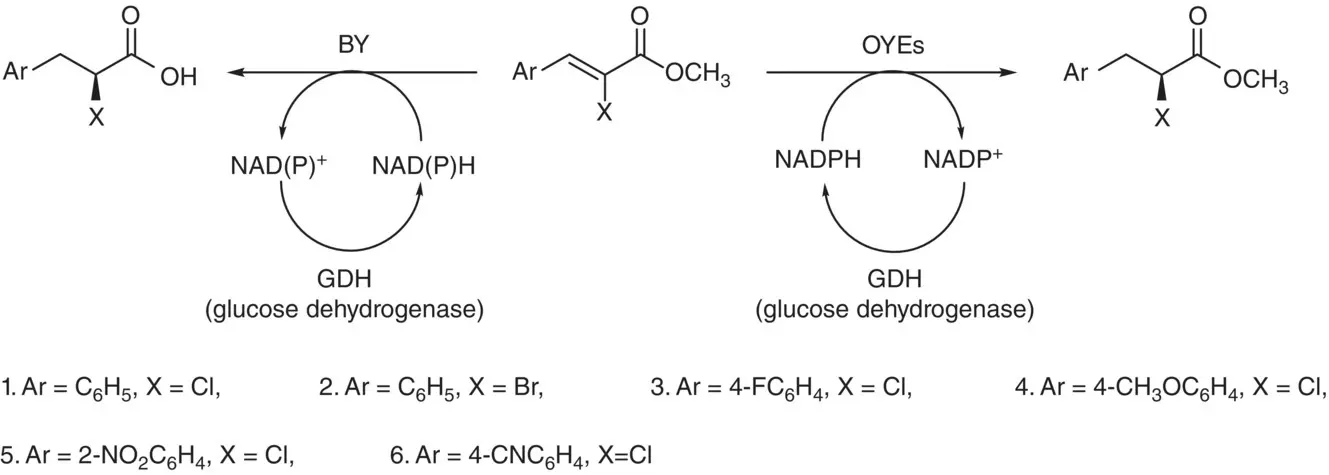
Scheme 2.43 BY fermentations and OYEs 1–3 mediated bioreductions of substrates 1–6.
The reduction of the double bond associated with α,β‐unsaturated ketones and catalyzed by microorganisms, animal, and plant cell cultures has long been studied and known by synthetic organic chemists. However, a strain of filamentous fungus identified as Aspergillus versicolor and named A. versicolor D‐1 was isolated and used for the reduction of the γ,δ‐double bond of the conjugated lactone in securinine to produce 14,15‐dihydrosecurinine with high stereospecificity ( Scheme 2.44) [186]. The NADPH‐dependent securinine reductase found in A. versicolor D‐1 is a cytosolic enzyme and is highly inducible in growing/resting cultures for securinine reduction. Moreover, it will not reduce the α,β‐double bond in 14,15‐dihydrosecurinine further. It was also found that the reductase is thermally unstable.
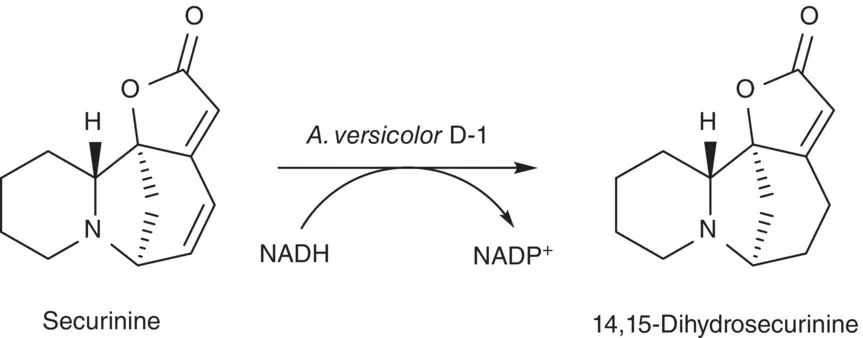
Scheme 2.44 The reduction of γ,δ‐double bond of the conjugated lactone in securinine.
References
1 1 Molinari, F. (2006). Curr. Org. Chem. 10: 1247–1263.
2 2 Bouonomenna, M.G. and Drioli, E. (2008). Org. Process Res. Dev. 12: 982–988.
3 3 Jia, A.Z., Lou, L.L., Zhang, C. et al. (2009). J. Mol. Catal. A 306: 123–129.
4 4 Celik, D., Bayraktar, E., and Mehmetoglu, D. (2004). Biochem. Eng. J. 17: 5–13.
5 5 Molinari, F., Gandolfi, R., Aragozzini, F. et al. (1999). Enzyme Microb. Technol. 25: 729–735.
6 6 Wu, J., Wang, J.‐L., Li, M.‐H. et al. (2010). Bioresour. Technol. 101: 8936–8941.
7 7 Orbegozo, T., Lavandera, I., Fabian, W.M.F. et al. (2009). Tetrahedron 65: 6805–6809.
8 8 Sagiroglu, A. and Yavuz, M.O. (2005). Artif. Cells Blood Substit. Biotechnol. 33: 343–355.
9 9 Norouzian, D., Akbarzadeh, A., Inanlou, D.N. et al. (2003). Enzyme Microb. Technol. 33: 150–153.
10 10 Carballeira Rodríguez, J.D., García‐Burgos, C., Quezada Alvarez, M.A. et al. (2004). Biotechnol. Bioeng. 87: 632–640.
11 11 Jia, X., Xu, Y., and Li, Z. (2011). ACS Catal. 1: 591–596.
12 12 Hilterhaus, L. and Liese, A. (2007). Adv. Biochem. Eng. Biotechnol. 105: 133–173.
13 13 Oedman, P., Wessjohann, L.A., and Bornscheuer, U.T. (2005). J. Org. Chem. 70: 9551–9555.
14 14 Hirscher, T., Gocke, D., Fernandez, M. et al. (2005). Tetrahedron 61: 7378–7383.
15 15 Scheid, G., Kuit, W., Ruijter, E. et al. (2004). Eur. J. Org. Chem. 2004: 1063–1074.
16 16 Nestl, B.M., Voss, C.V., Bodlenner, A. et al. (2007). Appl. Microbiol. Biotechnol. 76: 1001–1008.
17 17 Demir, A.S., Hamamci, H., Sesenoglu, O. et al. (2001). Tetrahedr. Asymm. 12: 1953–1956.
18 18 Smalleridge, A.J., Trewhalla, M.A., Maurice, A., and Wilkinson, A.K. (2003). Patent WO 2003018531 A1 20030306.
19 19 Rosche, B., Breuer, M., Hauer, B., and Rogers, P.L. (2004). Biotechnol. Bioeng. 86: 788–794.
20 20 Engel, S., Vyazmensky, M., Geresh, H. et al. (2003). Biotechnol. Bioeng. 83: 640–833.
21 21 Cheng, Y., Zhang, F., Rano, T.A. et al. (2002). Bioorg. Med. Chem. Lett. 12: 2419–2422.
22 22 Lunardi, I., Conceicao, G.J.A., Moran, P.J.S., and Rodrigues, J.A.R. (2005). Tetrahedr. Asymm. 16: 2515–2519.
23 23 Mitsukura, K., Sato, Y., Yoshida, T., and Nagasawa, T. (2004). Biotechnol. Lett. 26: 1643–1648.
24 24 Tanaka, M., Hirokane, Y., Mitsui, R., and Tsuno, T. (2001). J. Biosci. Bioeng. 91: 267–271.
25 25 Vicente, C., Fontaniella, B., Millanes, A.M. et al. (2003). Int. J. Cosmet. Sci. 25: 25–29.
26 26 Seshardri, R., Lamm, A.S., Khare, A., and Rosazza, J.P.N. (2008). Enzyme Microb. Technol. 43: 486–494.
27 27 Hamme, J.D., Singh, A., and Ward, O.P. (2003). Microbiol. Mol. Boil. Rev. 67: 503–549.
28 28 Beilen, J.B., Li, Z., Duetz, W.A. et al. (2003). Oil Gas Sci. Technol. 58: 427–440.
29 29 Labinger, J.A. (2004). J. Mol. Catal. A 220: 27–35.
30 30 Ayala, M. and Torres, E. (2004). Appl. Catal. A 272: 1–13.
31 31 van Beilen, J.B. and Funhoff, E.G. (2005). Curr. Opin. Biotechnol. 16: 308–314.
32 32 Bernhardt, R. (2006). J. Bacteriol. 124: 128–145.
33 33 Funhoff, E.G. and van Beilen, J.B. (2007). Biocatal. Biotransformation 25: 186–193.
34 34 Kawakami, N., Shoji, O., and Watanabe, Y. (2011). Angew. Chem. Int. Ed. 50: 5315–5318.
35 35 Weber, E., Seifert, A., Antonovici, M. et al. (2011). Chem. Commun. 47: 944–946.
36 36 Bordeaux, M., Galarneau, A., Fajula, F., and Drone, J. (2011). Angew. Chem. Int. Ed. 50: 2075–2079.
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Enzyme-Based Organic Synthesis»
Представляем Вашему вниманию похожие книги на «Enzyme-Based Organic Synthesis» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Enzyme-Based Organic Synthesis» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.












