The first big step forward for sequencing technology took place at Cambridge University, England, in the mid‐1950s in the laboratory of biologist Frederick Sanger. Well before gene sequences, in the earliest stages of our understanding of how genes function, Sanger discovered how to take a protein, break it down into its component parts, and, piecing the puzzle back together, determine the order of amino acids along a protein. His ingenious approach to understanding the sequencing of proteins eventually won him his first of two Nobel Prizes and was the conceptual precursor to contemporary DNA sequencing. (7) An understanding of proteins was also important because of the role these complex molecules play in an organism. Proteins receive their instructions from genes to carry out such diverse tasks as food digestion, production of energy in a cell, transmission of impulses in the nervous system, and the ability to smell, see, and hear. If genes and DNA are the material that perpetuate heredity and help determine an organism’s form and function, then proteins are the cell’s workhorses, carrying out the varied instructions inscribed in an individual’s DNA. Proteins can also play a harmful role in an organism. Genetic defects can cause the absence or overabundance of a particular protein, which in both cases can cause devastating illnesses. For example, phenylketonuria, or PKU, is a metabolic disease caused by a genetic defect that leaves individuals without a protein that breaks down the amino acid phenylalanine. A buildup of phenylalanine causes severe mental retardation. Babies diagnosed with the disease as part of newborn screening programs can have their diets altered to keep levels of phenylalanine low and avoid PKU’s dreadful effects. (8)
The method developed by Sanger exploited the chemistry of amino acids and proteins that had been well known for over 10 years. Just as nucleotides are the building blocks of DNA, amino acids are the building blocks of proteins. Sanger himself wrote in the journal Science :
In 1943 the basic principles of protein chemistry were firmly established. It was known that all proteins were built up from amino acid residues bound together by peptide bonds to form long polypeptide chains. Twenty different amino acids are found in most mammalian proteins, and by analytical procedures it was possible to say with reasonable accuracy how many residues of each one was present in a given protein. (9)
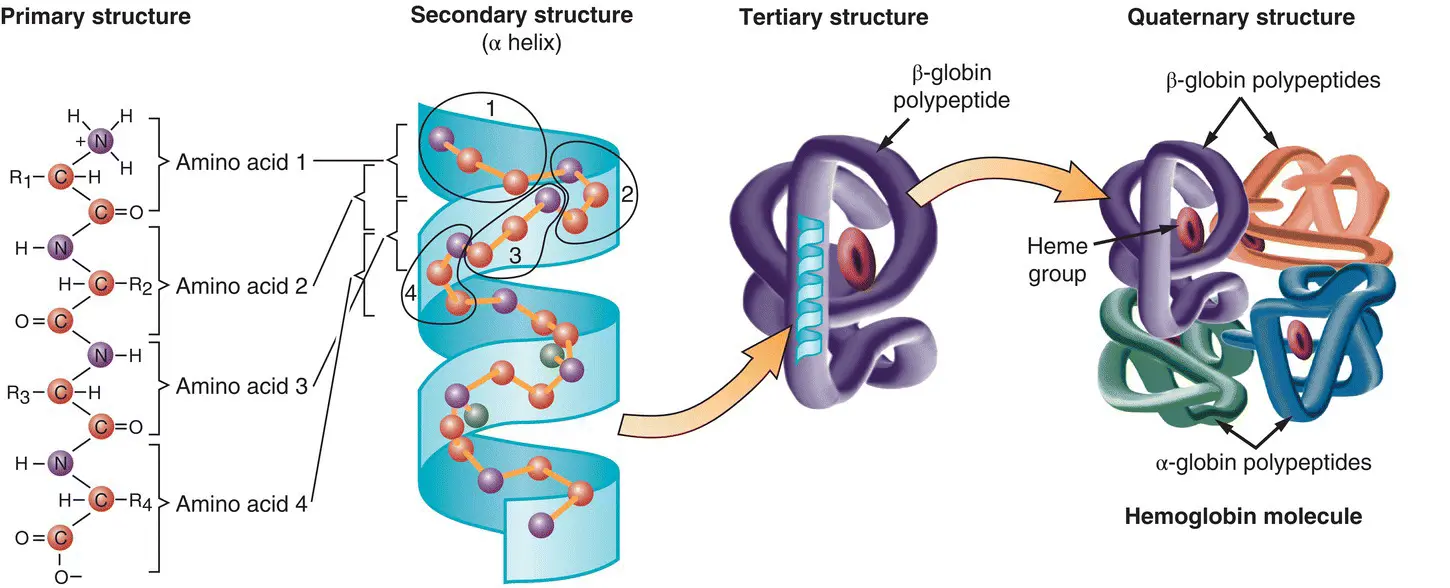
Figure 2.1This figure shows the way in which amino acids are the building blocks of proteins. In this case, we can see how a hemoglobin molecule is made up of a string of amino acids.
Credit: Wiley Publishers

Figure 2.2Frederick Sanger played a critical role in the development of molecular biology and in the technologies that enabled the sequencing of the human genome.
Credit: https://commons.wikimedia.org/wiki/File:Frederick_Sanger2.jpg
Sanger’s challenge was to figure out a way to read the order of the amino acids that determine a protein. For his experiments Sanger chose to use bovine, or cow, insulin because of its important medical significance and its relatively short length—only 105 amino acids. Sanger set out to find ways to read the unwieldy molecule, which by his method could be deciphered only by breaking the protein apart, looking at small stretches of four or five amino acids, and then conceptually putting the molecule back together like a puzzle to determine the full sequence.
Sanger determined that the exposure of insulin to certain chemicals could break the peptide bonds in a protein chain. Sanger was able to identify the kinds of amino acids these broken‐down parts contained. He then created groups of small chains of amino acids that could be “tiled,” or pieced together, to give a full‐length sequence of a protein. (10)
Sanger was considered to be “reticent, even shy, a man who worked with his hands, at the laboratory bench.” (11) Yet he also recognized the impact that his work would have on science and medicine.
In his address to the Nobel committee in 1958 Sanger underlined the importance of understanding the chemical nature of proteins. “These studies are aimed,” he said, “at determining the exact chemical structure of the many proteins that go to make up living matter and hence understand how these proteins perform their specific functions on which the processes of life depend.” He also hoped that his work “may reveal changes that take place in disease, and that our efforts may be of more practical use to humanity.” (12) This connection between proteins, genes, and medicine, uncovered in part by Sanger and his techniques, is at the heart of what lies ahead in genomics. Fred Sanger died at the age of 95 in 2013. His legacy is immense including an institute in the UK named after him and two Nobel Prizes. He was, as he said about himself, “a chap who messed about in his lab,” but he was also a chap who really made a difference to humankind. (13)
RESEARCH MILESTONE 2 : DECIPHERING THE GENETIC CODE
The most basic mechanisms and building blocks of heredity were, by the late 1950s, either solved or theoretically understood. But the link between genes and proteins was still not fully established. After all, nobody had yet explained exactly how DNA could produce a protein. The growing awareness that proteins were linear arrangements of amino acids and that genes were linear arrangements of nucleotides suggested to many scientists that this could mean only one thing—there was some code that connected the information in DNA to the production of proteins. But this was no simple code to crack, and scientists had been working on variations of this problem for at least a decade before the discovery of the structure of the double helix.
The intellectual spark that was a foundation for the solution of the DNA/protein code came from an unlikely source. Soon after the 1953 publication in Nature of their famous paper on the structure of DNA, Watson and Crick received a letter from George Gamow, a theoretical physicist and one of the architects of the big bang theory of the universe. Gamow’s letter sketched out an explanation for how an array of nucleic acids determined an array of amino acids. Gamow’s model, which detailed a list of 25 amino acids, turned out to be wrong. Paring down Gamow’s list to 20, Watson and Crick came up with the correct number of amino acids that make up proteins. (14) Over the next decade scientists conducted experiments that confirmed Watson and Crick’s list of amino acids and uncovered the DNA/protein coding scheme.
In DNA there are four linearly arranged nucleic acids (G, A, T, and C), whereas proteins are constructed from 20 linearly arranged amino acids. It was apparent from basic mathematics that the code was not based on a 1:1 relationship—the connection between DNA and proteins was not one nucleic acid to one amino acid (it would require at least 20 different nucleic acids to make a 1:1 ratio work). The code could also not be solved based on a 2:1 ratio. That is because there are only 16 ways G, A, T, and C can be arranged.
It turned out that the code is based on a 3:1 relationship and is therefore a series of nonoverlapping triplets of nucleic acids that code for single amino acids. Basic mathematics shows that there are 64 different ways to arrange four different bases in triplets. But there are only 20 types of amino acids. This is because some of the triplets, which are called codons, are redundant: they are just different ways to code for the same amino acid. Most amino acids have either two or four synonymous codons, although there are several exceptions. The amino acids methionine and tryptophan have no synonymous codons. Isoleucine has three, and serine, arginine, and leucine all have six.
Читать дальше














