1 ...7 8 9 11 12 13 ...37 In addition to the direct demonstration of a nucleic acid sequence by hybridization, amplification assays (the process of making additional copies of the specific sequence of interest) are of increasing importance in clinical microbiology. The most commonly used amplification assay is PCR ( Fig. 8). PCR uses a DNA polymerase that is stable at high temperatures that would denature and inactivate most enzymes. This thermostable DNA polymerase most often is isolated from the bacterium Thermus aquaticus . Its stability at high temperature enables the enzyme to be used without the need for replacement after the high-temperature conditions of the DNA denaturation step that occurs during each cycle of PCR:
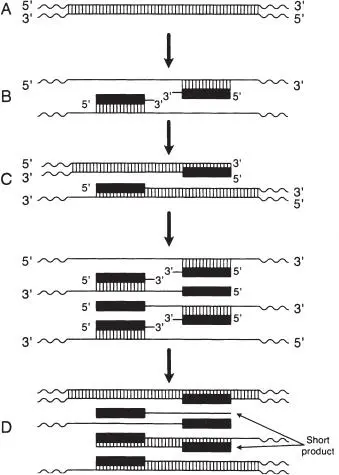
Figure 8 PCR. (A) In the first cycle, a double-stranded DNA target sequence is used as a template. (B) These two strands are separated by heat denaturation, and the synthetic oligonucleotide primers (solid bars) anneal to their respective recognition sequences in the 5′ → 3′ orientation. Note that the 3′ ends of the primers are facing each other. (C) A thermostable DNA polymerase initiates synthesis at the 3′ ends of the primers. Extension of the primer via DNA synthesis results in new primer-binding sites. The net result after one round of synthesis is two “ragged” copies of the original target DNA molecule. (D) In the second cycle, each of the four DNA strands in panel C anneals to primers (present in excess) to initiate a new round of DNA synthesis. Of the eight single-stranded products, two are of a length defined by the distance between and including the primer-annealing sites; these “short products” accumulate exponentially in subsequent cycles. (Reprinted from Manual of Clinical Microbiology , 7th ed, ©1999 ASM Press, with permission.)
1 1. The target DNA sequence is heated to a high temperature that causes the double-stranded DNA to denature into single strands.
2 2. An annealing step follows, at a lower temperature than the denaturation step above, during which sets of primers, with sequences designed specifically for the PCR target sequences, bind to these target sequences.
3 3. Last is an extension step, during which the DNA polymerase completes the target sequence between the two primers.
Assuming 100% efficiency, the above three steps generate two copies of the target sequence. Multiple cycles (such as 30) in a thermal cycler result in a tremendous amplification of the number of sequences, so that the sequence is readily detectable using any of a variety of methods—gel electrophoretic, colorimetric, chemiluminescent, or fluorescent.
When the specific target nucleic acid is RNA rather than DNA, a cDNA sequence is made with the enzyme reverse transcriptase (RT) before PCR amplification in a procedure known as RT-PCR. Examples of pathogens for which RT-PCR is used include the RNA-containing viruses HIV-1 and hepatitis C virus (HCV).
An additional feature of PCR is that the amplified nucleic acid products can be directly sequenced. These sequences can be compared with sequences found in publicly accessible databases. This allows, for example, the identification of a bacterial organism to the level of species on the basis of a sequence of hundreds of bases in the rRNA or, if the sequence is less closely related to sequences within the database, to the level of genus. In some cases, the organism may be an entirely new one. This method of PCR and sequencing of the product for the purposes of bacterial identification is being used in clinical microbiology for the identification of slow-growing or difficult-to-identify organisms such as Mycobacterium spp., Nocardia spp., and anaerobic organisms. However, mass spectrometry has recently entered clinical microbiology and will likely replace ribosomal gene sequencing as the method of choice for these organisms, as well as all other bacteria and fungi. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) allows the identification of organisms by their protein spectra. Although initial instrumentation is expensive, identifications can be performed for less than $1 and in at little as 20 minutes. Many clinical laboratories are already using MALDI-TOF as the primary method for identifying bacteria.
After the invention of PCR, a number of other amplification assays were developed, some of which have entered the clinical microbiology laboratory. Transcription-mediated amplification (TMA), which does not require a thermal cycler, relies on the formation of cDNA from a target single-stranded RNA sequence, the destruction of the RNA in the RNA-DNA hybrid by RNase H, and the formation of double-stranded cDNA (which can serve as transcription templates for T7 RNA polymerase). A similar procedure occurs during the nucleic acid sequence-based amplification (NASBA) assay. Strand-displacement amplification (SDA) does not require a thermal cycler and has two phases in its cycle: a target generation phase during which a double-stranded DNA sequence is heat denatured, resulting in two single-stranded DNA copies; and an exponential amplification phase in which a specific primer binds to each strand at the cDNA sequence. DNA polymerase extends the primer, forming a double-stranded DNA segment that contains a specific restriction endonuclease recognition site, to which a restriction enzyme binds, cleaving one strand of the double-stranded sequence and forming a nick, followed by extension and displacement of new DNA strands by DNA polymerase.
All of these assays—PCR, TMA/NASBA, and SDA—have one thing in common: they amplify the target nucleic acid sequence, making many, many copies of the sequence. As you might imagine, there is the possibility that small quantities of the billions of amplified target nucleic acid sequences can contaminate a sample that will then undergo amplification testing, resulting in false-positive results. Steps are taken to minimize contamination, including physical separation of specimen preparation and amplification areas, positive displacement pipettes, and both enzymatic (in PCR) and nonenzymatic methods to destroy the amplified products.
An alternative method of demonstrating the presence of a specific nucleic acid sequence that does not require the amplification of the target is by amplification of the signal. One example is branched DNA (bDNA) technology ( Fig. 9), which is used particularly in quantitative assays, such as HIV and HCV viral load determinations. In this assay, specific oligonucleotides hybridize to the sequence of interest and capture it onto a solid surface. In addition, a set of synthetic enzyme-conjugated branched oligonucleotides hybridize to the target sequence. When an appropriate substrate is added, light emission is measured and compared with a standard curve. This permits quantitation of the target sequence. As there is no amplified sequence to be concerned about, the risk of contamination is dramatically reduced. Another example that is widely used is a hybrid capture test for human papillomavirus (HPV) detection. In this test, HPV DNA is denatured and bound to complementary RNA probes. This hybrid is then “captured” by immobilized anti-hybrid antibodies. A chemiluminescent reaction allows for the detection of DNA-RNA hybrids and therefore HPV DNA in the sample.
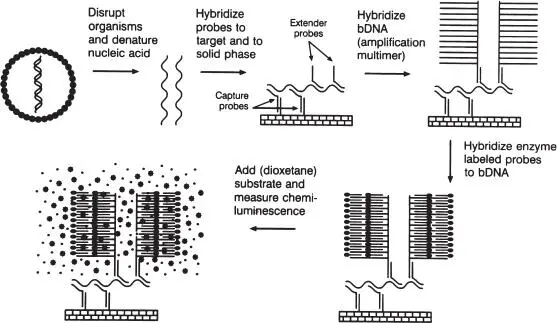
Figure 9 bDNA-based signal amplification. Target nucleic acid is released by disruption and is captured onto a solid surface via multiple contiguous capture probes. Contiguous extended probes hybridize with adjacent target sequences and contain additional sequences homologous to the branched amplification multimer. Enzyme-labeled oligonucleotides bind to the bDNA by homologous base pairing, and the enzyme-probe complex is measured by detection of chemiluminescence. All hybridization reactions occur simultaneously. (Reprinted from Manual of Clinical Microbiology , 7th ed, ©1999 ASM Press, with permission.)
Читать дальше














