Transcriptional Regulation
Expression of a gene is often regulated by controlling the amount of mRNA that is made from the gene. This is called transcriptional regulation. It makes sense to regulate gene expression at this level, as it is wasteful to make mRNAs if the expression of the gene is going to be inhibited at a later stage. Also, bacterial genes are often arranged in a polycistronic unit, or operon; if the genes in this unit are involved in a related function, they can all be regulated simultaneously by regulating the synthesis of the polycistronic mRNA of that operon.
Regulation of transcription of an operon usually occurs at the initial stages of transcription, at the promoter. Whether or not a gene is expressed depends on whether the promoter for the gene is used to make mRNA. Transcriptional regulation at the promoter for a gene can be determined by specific recognition of the promoter by RNA polymerase holoenzyme containing an alternative sigma factor. Regulation can also use regulatory proteins that can act either negatively or positively, depending on whether the regulatory gene product is a transcriptional repressoror a transcriptional activator, respectively. The difference between regulation of transcription by repressors and activators is illustrated in Figure 2.43. A repressor binds to the DNA at an operatorsequence close to, or even overlapping, the promoter and prevents RNA polymerase from using the promoter, often by physically obstructing access to the promoter by the RNA polymerase. An activator, in contrast, usually binds upstream of the promoter at an activator site, where it can help the RNA polymerase bind to the promoter or help open the promoter after the RNA polymerase binds. Sometimes, a transcriptional regulator can be a repressor on some promoters and an activator on other promoters, depending on where it binds relative to the start site of transcription.
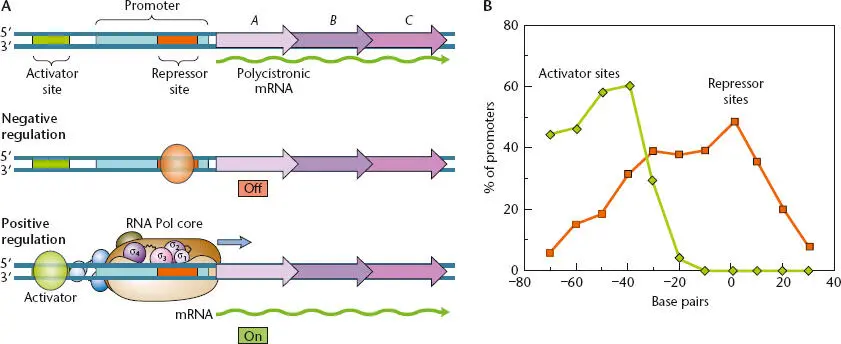
Figure 2.43 (A)The two general types of transcriptional regulation. In negative regulation, a repressor binds to a repressor-binding site (or operator) and turns expression of the operon off. In positive regulation, an activator protein binds upstream of the promoter and turns expression of the operon on. (B)Graph showing the most common locations of activator sites relative to repressor sites. Activator sites are usually farther upstream. Each data point indicates the middle of the known region on the DNA where a regulatory protein binds. Zero on the x axis marks the start point of transcription. Modified from Collado-Vides J, Magasanik B, Gralla JD, Microbiol Rev 55:371–394, 1991.
The activity of a regulatory protein can itself be modified by the binding of small molecules called effectorswhich affect its activity (note that the term effector is also used for proteins that are translocated by pathogens into eukaryotic cells). Effectors used in gene regulation are often molecules that can be used by the cell if the regulated operon is expressed or essential metabolites that do not have to be made by the cell if their concentration is already high. If the small-molecule effector causes transcription of the operon to be turned on (for example, by binding to a repressor and changing its structure so that the repressor can no longer bind to the DNA), the small molecule is called an inducer. If binding of the effector to a repressor causes the operon to be turned off, the small molecule is called a corepressor. The activity of regulatory proteins can also be modulated by posttranslational modification (e.g., phosphorylation [see Box 12.3]) or by interaction with other proteins or RNAs.
Not all transcriptional regulation occurs at the promoter, however. Sometimes transcription starts and then stops prematurely, resulting in synthesis of a truncated mRNA that does not include the protein-coding sequence. Such regulation is called attenuation of transcription. These and other mechanisms of transcriptional regulation are discussed in subsequent chapters.
Posttranscriptional Regulation
Expression of a gene can be regulated at later stages in gene expression (see chapter 11). For example, the mRNA may be degraded by RNases as soon as it is made, before it can be translated. In translational regulation, translation of the mRNA can be regulated to determine how much of the protein product is made. Posttranslational regulationresults in regulation of the activity of a protein product of a gene by degradation by proteases or modifications, such as phosphorylation or methylation, depending on the conditions in which the cell finds itself. In addition, the product of a pathway may inhibit the activity of an enzyme in the pathway by a process called feedback inhibition. In general, a type of regulation of gene expression that operates after the mRNA for a gene has been made is called posttranscriptional regulation. Specific examples of posttranscriptional regulation are also discussed in subsequent chapters.
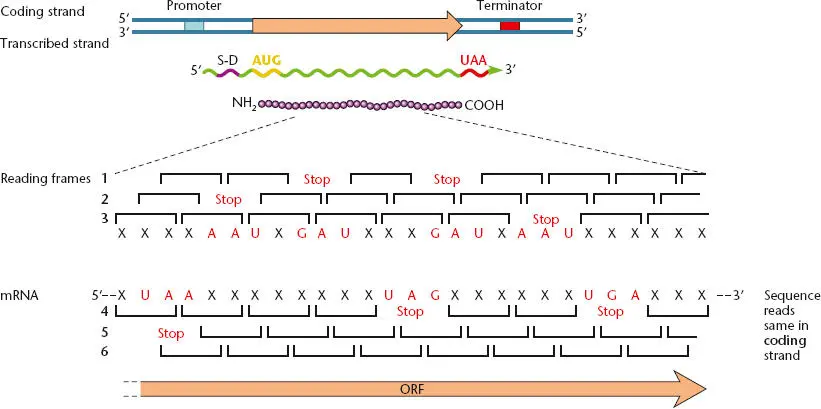
Figure 2.44 Relationship between gene structure in DNA and the coding sequence in mRNA. Each of the different reading frames in the two strands of DNA may contain open reading frames (ORFs), but generally, only one ORF in each region (reading frame 6 in the diagram) is translated to yield a polypeptide product.
We have introduced a lot of detail in this chapter, so it is worth reviewing some of the most important concepts and terms. As with any field, molecular genetics has its own jargon, and in order to follow a paper or seminar that includes some molecular genetics, familiarity with this jargon is very helpful.
Figure 2.44shows a typical gene with a promoter and transcription terminator. The mRNA is transcribed beginning at the promoter and ending at the transcription terminator. The direction on the DNA or RNA is indicated by the direction of the phosphate bonds between the carbons on the ribose or deoxyribose sugars in the backbone of the polynucleotide. These carbons are labeled with a prime to distinguish them from the carbons in the bases of the nucleotides. On one end of the RNA, the 5′ carbon of the terminal nucleotide is not joined to another nucleotide by a phosphate bond. Therefore, this is called the 5′ end. Similarly, the other end is called the 3′ end, because the 3′ carbon of the last nucleotide on this end is not joined to another nucleotide by a phosphate bond. The direction on DNA or RNA from the 5′ end to the 3′ end is called the 5′-to-3′ direction. An RNA polymerase molecule synthesizes mRNA in the 5′-to-3′ direction, moving 3′ to 5′ on the transcribed strand (or template strand) of DNA. The opposite strand of DNA from the transcribed strand has the same sequence and 5′-to-3′ polarity as the RNA, so it is called the coding strand (or nontemplate strand). Sequences of DNA in the region of a gene are usually shown as the sequence of the coding strand. A sequence that is located in the 5′ direction of another sequence on the coding strand is upstream of that sequence, while a sequence in the 3′ direction is downstream. Therefore, the promoter for a gene and the S-D sequences are both upstream of the initiation codon, while the termination codon and the transcription termination sites are both downstream.
Читать дальше













