Finally, yeasts condense acetic acid (in the form of acetyl‐CoA) and pyruvic acid to produce citramalic acid (0–300 mg/l) and dimethylglyceric acid (0–600 mg/l) ( Figure 2.18). These compounds have little sensory impact.
2.3.6 Degradation of Malic Acid by Yeast
Saccharomyces cerevisiae partially degrades malic acid (10–25%) in the must during alcoholic fermentation. Different strains degrade varying amounts of this acid and degradation is more significant when the pH is low. Alcoholic fermentation is the main pathway degrading malic acid. The pyruvic acid resulting from this transformation is decarboxylated into acetaldehyde, which is then reduced to ethanol. Malic enzyme is responsible for the transformation of malic acid into pyruvic acid ( Figure 2.19). This oxidative decarboxylation requires NAD +(Fuck and Radler, 1972). This malo‐alcoholic fermentation lowers wine acidity significantly more than malolactic fermentation does.
Schizosaccharomyces differs from wine yeasts. The alcoholic fermentation of malic acid is complete in yeasts of this genus, which possess an active malate transport system. In S. cerevisiae , malic acid penetrates the cell by simple diffusion. Yet at present no attempts to use Schizosaccharomyces in winemaking to break down the malic acid in musts have been successful (Peynaud et al ., 1964; Carre et al ., 1983). First of all, the implantation of these yeasts in the presence of S. cerevisiae is difficult in a non‐sterilized must. Secondly, their optimum growth temperature (30°C), higher than for S. cerevisiae , imposes warmer fermentation conditions. Sometimes, the higher temperature adversely affects the sensory quality of wine. Finally, some grape varieties fermented by Schizosaccharomyces do not express their varietal aromas. The acidic Gros Manseng variety produces a very fruity wine when correctly vinified with S. cerevisiae , but has no varietal aroma when fermented by Schizosaccharomyces . To resolve these problems, some researchers have used non‐proliferating populations of Schizosaccharomyces enclosed in alginate balls. These populations degrade the malic acid in wines having already completed their alcoholic fermentation (Magyar and Panyik, 1989; Taillandier and Strehaiano, 1990). Although no sensory defect is found in these wines, the techniques have not yet been developed for practical use.
Today, molecular biology permits another strategy for making use of the ability of Schizosaccharomyces to ferment malic acid. It consists of integrating Schizosaccharomyces malate permease genes and the malic enzyme ( Mae 1 and Mae 2) in the S. cerevisiae genome (Van Vuuren et al ., 1996). The technological interest of a wine yeast genetically modified in this manner is not yet clear, nor are the risks of its proliferation in wineries and nature.
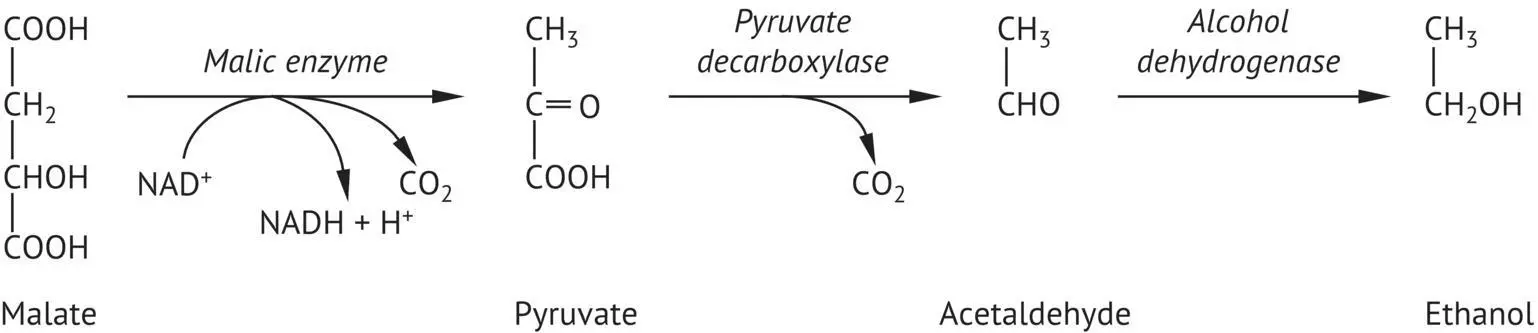
FIGURE 2.19 Decomposition of malic acid by yeasts during alcoholic fermentation.
2.4 Metabolism of Nitrogen Compounds
The nitrogen requirements of wine yeasts and the nitrogen supply in grape musts are discussed later ( Section 3.4.2). The following section covers the general mechanisms of assimilation, biosynthesis, and degradation of amino acids in yeasts. The consequences of these metabolisms, which occur during alcoholic fermentation and affect the production of higher alcohols and their associated esters in wine, are also discussed.
2.4.1 Amino Acid Synthesis Pathways
The ammonium ion and amino acids found in grape must supply the yeast with nitrogen. The yeast can also synthesize most of the amino acids necessary for constructing its proteins. It fixes an ammonium ion on a carbon skeleton derived from the metabolism of sugars. The yeast uses the same reaction pathways as all organisms. Glutamate and glutamine play an important role in this process (Cooper, 1982; Magasanik, 1992).
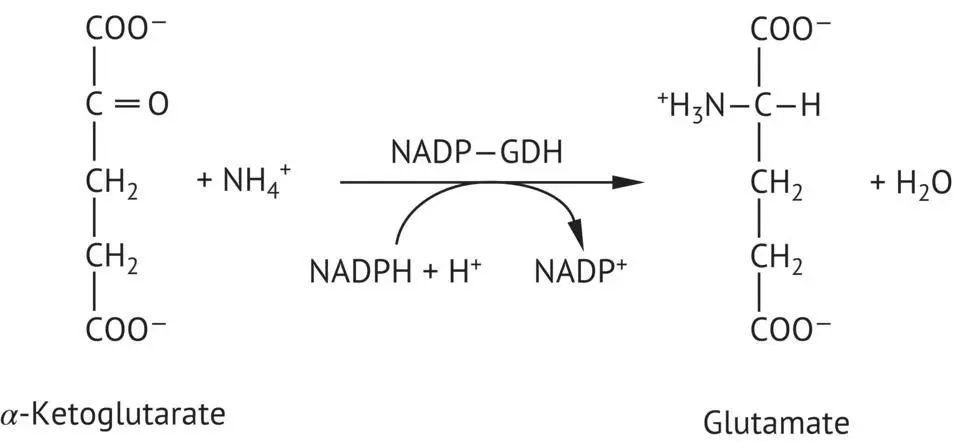
FIGURE 2.20 Incorporation of the ammonium ion in α ‐ketoglutarate catalyzed by NADP glutamate dehydrogenase (NADP‐GDH).
NADP +glutamate dehydrogenase (NADP +‐GDH), a product of the GDH1 gene, produces glutamate ( Figure 2.20) from an ammonium ion and an α ‐ketoglutarate molecule. The latter is an intermediate product of the citric acid cycle. The yeast also possesses an NAD +glutamate dehydrogenase (NAD +‐GDH), produced by the GDH2 gene. This dehydrogenase is involved in the oxidative catabolism of glutamate. It produces the inverse reaction of the previous one, liberating the ammonium ion used in the synthesis of glutamine. NADP‐GDH activity is at its maximum when the yeast is cultivated on a medium containing exclusively ammonium as its source of nitrogen. The NAD‐GDH activity, however, is at its highest level when the principal source of nitrogen is glutamate. Glutamine synthetase (GS) produces glutamine from glutamate and ammonium. This amidationrequires the hydrolysis of an ATP molecule ( Figure 2.21).
Through transamination reactions, glutamate then serves as an amino group donor in the biosynthesis of different amino acids. Pyridoxal phosphate (PLP) is the transaminase cofactor ( Figure 2.22); it is derived from pyridoxine (vitamin B 6).
The carbon skeleton of amino acids originates from intermediates of glycolysis (pyruvate, 3‐phosphoglycerate, and phosphoenolpyruvate), the citric acid cycle ( α ‐ketoglutarate and oxaloacetate), or the pentose phosphate cycle (ribose 5‐phosphate and erythrose 4‐phosphate). Some of these reactions are very simple, such as the formation of aspartate or alanine by transamination of glutamate into oxaloacetate or pyruvate:


Other biosynthetic pathways are more complex, but still occur in yeasts as in the rest of the living world. The amino acids can be classified into six biosynthetic families, depending on their nature and their carbon precursor ( Figure 2.23):
1 In addition to glutamate and glutamine, proline and arginine are formed from α‐ketoglutarate.
2 Asparagine, methionine, lysine, threonine, and isoleucine are derived from aspartate, which comes from oxaloacetate. ATP can activate methionine to formS‐adenosylmethionine, which can be demethylated to form S‐adenosylhomocysteine, the hydrolysis of which liberates adenine to produce homocysteine. FIGURE 2.21 Amidation of glutamate into glutamine by glutamine synthetase (GS). FIGURE 2.22 Pyridoxal phosphate (PLP) and pyridoxamine phosphate (PMP). FIGURE 2.23 General biosynthesis pathways of amino acids.
3 Pyruvate is the starting point for the synthesis of alanine, valine, and leucine.
4 3‐Phosphoglycerate leads to the formation of serine and glycine. The condensation of homocysteine and serine produces cystathionine, a precursor of cysteine.
5 The imidazole ring of histidine is formed from ribose 5‐phosphate and adenine coming from ATP.
Читать дальше
















