Pathology of Genetically Engineered and Other Mutant Mice
Здесь есть возможность читать онлайн «Pathology of Genetically Engineered and Other Mutant Mice» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Pathology of Genetically Engineered and Other Mutant Mice
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:3 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 60
- 1
- 2
- 3
- 4
- 5
Pathology of Genetically Engineered and Other Mutant Mice: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Pathology of Genetically Engineered and Other Mutant Mice»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
An updated and comprehensive reference to pathology in every organ system in genetically modified mice Pathology of Genetically Engineered and Other Mutant Mice
Pathology of Genetically Engineered and Other Mutant Mice
Pathology of Genetically Engineered and Other Mutant Mice — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Pathology of Genetically Engineered and Other Mutant Mice», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
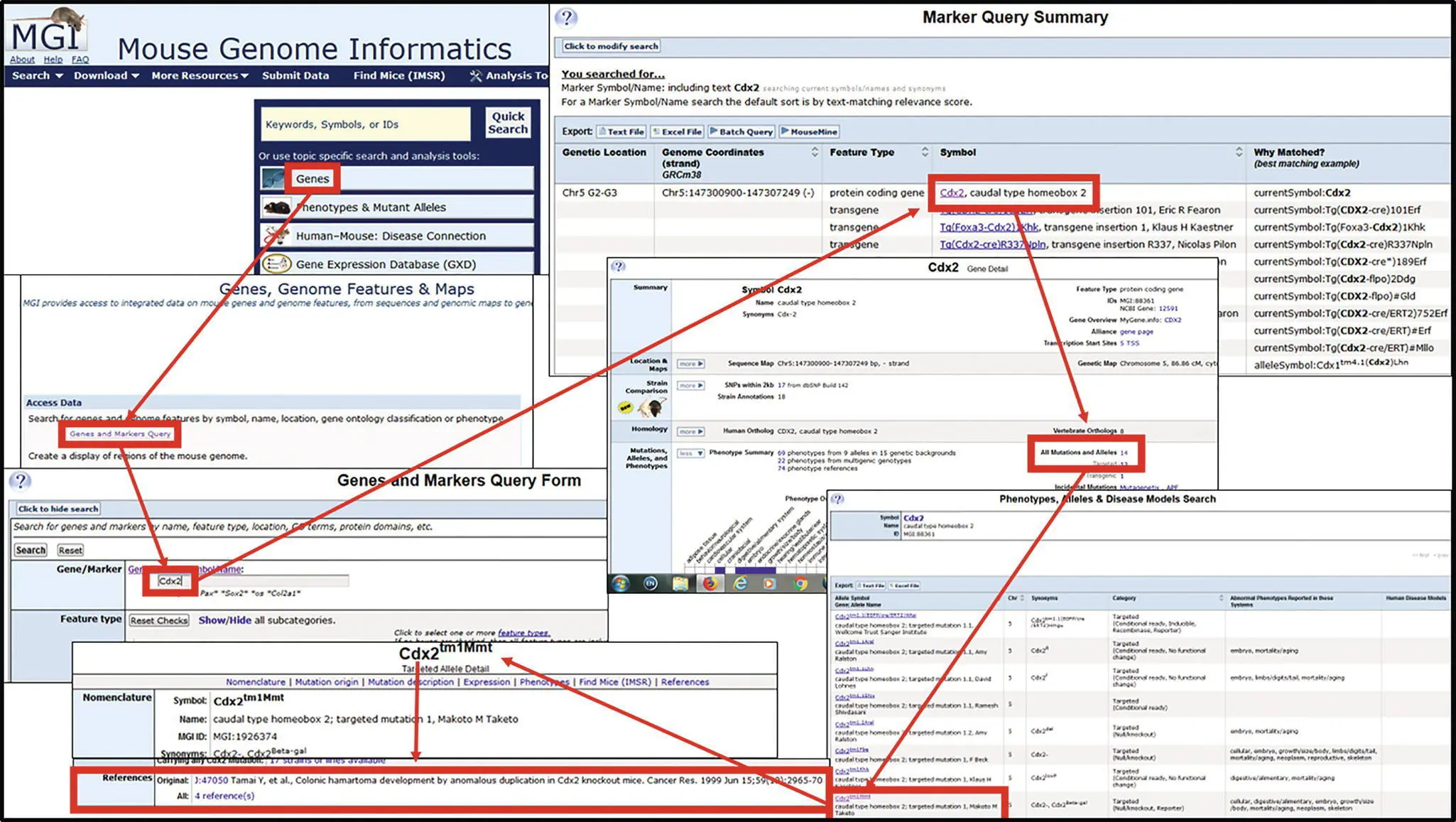
Figure 3.2 Finding the gene and allele symbol on Mouse Genome Informatics.One of the chapter authors inquired about how to find the correct allele symbol to use relative to a specific reference
(Source: Tamai Y, Nakajima R, Ishikawa T, Takaku K, Seldin MF, Taketo MM. Colonic hamartoma development by anomalous duplication in Cdx2 knockout mice. Cancer Res. 1999 Jun 15;59(12):2965‐70).
Go to the Mouse Genome Informatics home page ( http://www.informatics.jax.org/) and select “Gene.” The link goes to “Genes, Genome Features & Maps.” Then select “Genes and Marker Query.” In the Gene/Marker query box, enter the gene symbol or mutant mouse name, in this case Cdx2 was entered. Note, while gene symbols should be in italics no italics are given on this website or many other websites. This will give you all the genes for which this is part of the symbol or where Cdx2 was used as a synonym at some point in time. Selecting Cdx2 will yield the “Gene Detail” page. In the lower right area is the link to find all 14 of the currently reported allelic mutations (as of 1 May 2020) or the number of specific types of mutations. By selecting “All Mutations and Alleles” a summary field will appear. But which one is correct? By looking at the last author on the referenceit is obvious from the summary. This is confirmed by selecting “ Cdx2 tm1Mmt” to reveal the data sheet on this specific allelic mutation. At the bottom of the field is the reference that is the original reference provided.
Source: The Jackson Laboratory.
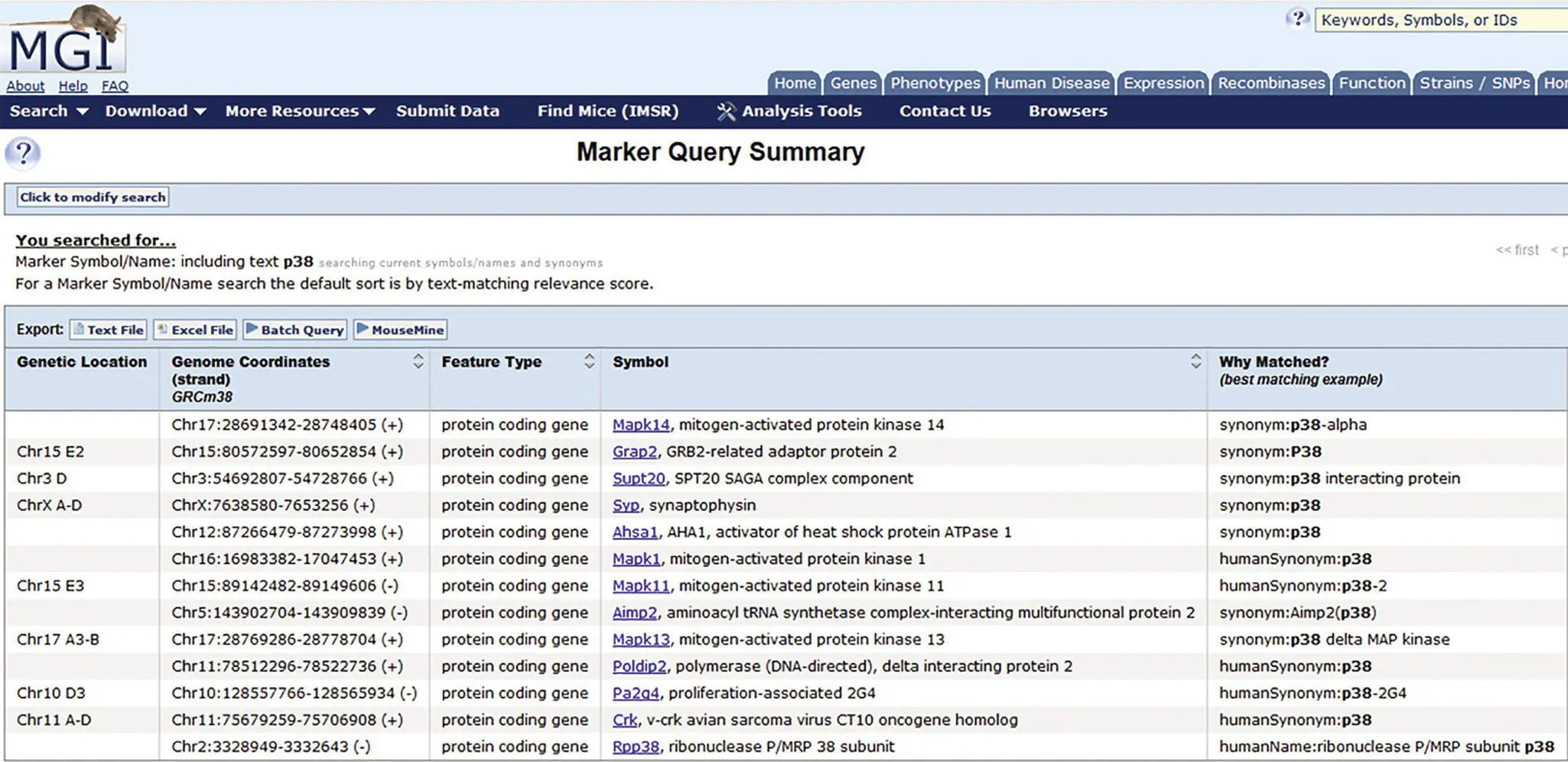
Figure 3.3 Current symbols assigned to a variety of unrelated genes originally published as p38.If one searches for the gene symbol p38 the results indicate numerous genes on different chromosomes that at one time were all called p38 . The new names not only separate out the different genes but also reflect to a degree what is currently known about the gene function. By following the links you can obtain much more detail on each gene, the number and details on allelic mutations, and links to the original and many subsequent publications on the topic (MGI accessed 22 April 2020).
Table 3.3 Inbred strains of mice.
| Advantages Genetic and phenotypic uniformity (smaller numbers needed)Well characterized (pathology and physiology)For most standard inbred strains >200 generationsIdeal controls (both biological and sequencing)Permits clear genetic mappingEnables identification of modifier genes |
| Disadvantages Not as robust (smaller, lower reproductive performance [fecundity], shorter lifespan)Strain‐specific characteristics (deleterious mutations causing strain specific diseases)Expensive (when difficult to maintain) |
| Uses Widely used in all types of researchModels for human diseaseBackground for mice with spontaneous and induced mutations |
Table 3.4 Inbred strain nomenclature.
| C57BL/6J or C57BL/10J |
| C57BLis the parental strain name (since C57BL/6J is the actual strain name) |
| 6or 10indicate the substrain line number (C57BL/10J are prone to serious heart disease not seen in C57BL/6J) [8] |
| Jindicates the breeder (The Jackson Laboratory) of these substrains |
| C3H/HeJ or C3H/HeN |
| C3His the strain generated by Strong from a cross of Bagg albino with DBA |
| Heis the substrain from the W.E. Heston laboratory at the National Cancer Institute |
| Jindicates the subline received from Heston then bred by The Jackson Laboratory |
| Nindicates the substrain bred at the National Institutes of Health |
An inbred strain is designated in capital letters.
A substrain is identified using a forward slash (/) after the strain name followed by a lab code of the strain breeder.
Further substrains derived from the founder add to the end of the lab code of subsequent breeders without another forward slash.
Note that C3H/HeJ has a mutation in the Tlr4 gene making it highly susceptible to gram negative bacterial infection while the C3H/HeN substrain is wildtype for Tlr4 .
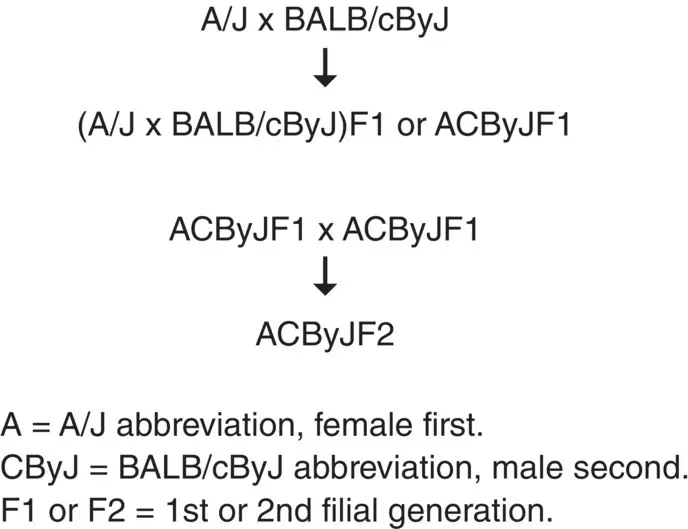
Figure 3.4 Nomenclature for hybrid stocks.
Table 3.5 Examples of inbred strain abbreviations.
| 129P3/J = 129P3 |
| 129S1/SvImJ = 129S1 |
| A/HeJ = AHe |
| A/J = A |
| AKR/J = AK |
| BALB/cByJ = CByJ |
| BALB/cJ = C |
| C57BL = B |
| C57BL/6J = B6 |
| C57BL/6JEi = B6Ei |
| C57BL/6NJ = B6NJ |
| C57BL/10J = B10 |
| C57BR/cdJ = BR |
| C57L = L |
| CBA/CaJ = CBACa |
| CBA/J = CBA |
| C3H/HeJ = C3 |
| C3HeB/FeJ = C3Fe |
| DBA/1J = D1 |
| DBA/2J = D2 |
| NZB/BINJ = NZB |
| NZW/LacJ = NZW |
| RIIIS/J = R3 |
| SJL/J = SJL or J |
| SWR/J = SW |
Table 3.6 F1 and F2 hybrid mice.
| F1 hybrids | F2 hybrids |
|---|---|
| Advantages Genetic and phenotypic uniformityContains a 50 : 50 mix of both parental strainsHybrid vigorAccepts transplants of tissue from mice of either parental strain | Advantages Hybrid vigorF2 hybrids do not have 50 : 50 mix but do have some mice in the population homozygous for some alleles and are valuable as a population for mutant gene mapping |
| Disadvantage Not self‐perpetuating | Disadvantages Not self‐perpetuatingGenotype and phenotype NOT uniformParental strains most likely reject tissue transplants |
| Uses Radiation researchBehavioral researchBioassays of nutrients, drugs, pathogens, or hormonesTransplant recipientsBackground for transgenics and some deleterious mutations | Use Control for many targeted (“knockout”) mutations on a mixed B6; 129 |
Sibling breeding F1 hybrid mice for one generation produces an F2 hybrid population ( Figure 3.4), and continued inbreeding after that eventually leads to the creation of recombinant inbred strains (see below). The F2 hybrid population also has hybrid vigor, but is distinct from the F1 hybrid population because meiosis has resulted in recombinations between the parental chromosomes such that there may be some homozygosity in a minority of the segregating alleles. This makes an F2 hybrid population a tremendous tool for mapping, especially for mapping recessive mutations. For example, if an interesting phenotype is observed in an inbred strain, such as an abnormal hair coat, hair interior defect ( hid ) in all AKR/J mice, these animals can be crossed with another inbred strain, such as BALB/cByJ, that has normal hair ( Figure 3.5). As hid in this example is an autosomal recessive mutation, the obligate heterozygous F1 progeny of this cross will all have a normal hair coat. When these heterozygous progeny are intercrossed to create F2 hybrids, 25% of these mice are expected to have the abnormal hair phenotype. The genetic interval carrying the mutant sequence derives from AKR/J so by screening the DNA from the mutant and unaffected mice in the population with molecular markers distributed evenly across all chromosomes, a pattern emerges in which markers specific to AKR/J and not BALB/cByJ skew to increased homozygosity in the region of the genome specific to the mutation. Usually this works relatively quickly to identify the region in which the mutated gene is located. However, in this particular case, less than 15% of the F2 mice were affected ( Table 3.7) due to modifier genes in BALB/cByJ. Similar crosses with other strains resulted in the expected 25% distribution. This illustrates two points, the deviation from Mendelian expectation due to modifiers and the value of doing mapping crosses with different inbred strains, which in this instance identified different genetic intervals that together reduced the interval and the number of candidate genes ( Figure 3.6) [9, 10].
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Pathology of Genetically Engineered and Other Mutant Mice»
Представляем Вашему вниманию похожие книги на «Pathology of Genetically Engineered and Other Mutant Mice» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Pathology of Genetically Engineered and Other Mutant Mice» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.












