Pathology of Genetically Engineered and Other Mutant Mice
Здесь есть возможность читать онлайн «Pathology of Genetically Engineered and Other Mutant Mice» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Pathology of Genetically Engineered and Other Mutant Mice
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:3 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 60
- 1
- 2
- 3
- 4
- 5
Pathology of Genetically Engineered and Other Mutant Mice: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Pathology of Genetically Engineered and Other Mutant Mice»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
An updated and comprehensive reference to pathology in every organ system in genetically modified mice Pathology of Genetically Engineered and Other Mutant Mice
Pathology of Genetically Engineered and Other Mutant Mice
Pathology of Genetically Engineered and Other Mutant Mice — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Pathology of Genetically Engineered and Other Mutant Mice», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
Collaborative Cross (CC) Mice
Another variation of RI lines is the Collaborative Cross. This is a complex set of crosses using eight different inbred strains that represent a great deal of genetic diversity [17, 18]. The goal was to create over 1000 new inbred strains. Due to reproductive failure and low viability of many strains, the current goal is to create 200 strains. These mice present with great phenotypic variability, a reflection of the great genetic diversity. These can be used to screen for specific phenotypes [19, 20]. When a large enough cohort are examined, phenotyped, and genotyped using the Mouse Universal Genotyping Array (Single Nucleotide Polymorphism [SNP] Array), it is possible to identify candidate gene regions and even the gene(s) responsible for the phenotype [21].
Collaborative Cross mice are designated by CC for Collaborative Cross, the line number, forward slash, and the lab that created it followed by the laboratory that currently maintains it. For example, CC001/UncJ (stock number 021238) is line number one, generated at the University of North Carolina, and currently maintained and distributed by The Jackson Laboratory.
Congenic Mice
When a genetic mutation arises spontaneously or is created in a specific mouse strain and the investigator wants to characterize it on another inbred strain, this can be accomplished by a modification of inbreeding to create what is called a congenic strain ( Table 3.9). For example, if you have an albino (white) strain carrying a single gene mutation, specifically a recessive mutation such as hairless on the HRS/J inbred strain (donor strain), and want to move it onto a predominantly C57BL/6J background (host strain), one parent from each strain is mated together to create F1 hybrids (see below). These F1 progeny are intercrossed to create F2 mice, ideally but not always 25% of which will be homozygous for the recessive hairless mutant allele. Only the mutant mice are then crossed back (backcrossed) to C57BL/6J mice to produce the N2 generation. This is repeated 10 times (N10) to create a fully congenic mouse strain [22, 23], with congenic nomenclature permitted to be used at N5. At N10, the strain will be mathematically >99% host background. Unfortunately, there will always be some of the parental strain DNA flanking the gene selected for, known as the congenic interval, and the possibility of trace amounts of donor sequence elsewhere in the genome. The more backcrosses to B6, the smaller this congenic interval should become and the lower the likelihood of traces of donor sequence but the genetic background will never entirely become B6. Creating a group of congenic strains on the same host background controls for phenotypic variability caused by genetic background in comparative or combinatorial assessments of mutation‐induced phenotypes.
This description is only true for recessive mutations that cannot be genotyped and even in those instances, a handful of researchers might go straight to N2 before the intercross, especially if it is a homozygote that cannot be used to breed. Dominant mutations or alleles that can be genotyped do not require the intercross step so can be repeatedly backcrossed at each generation.
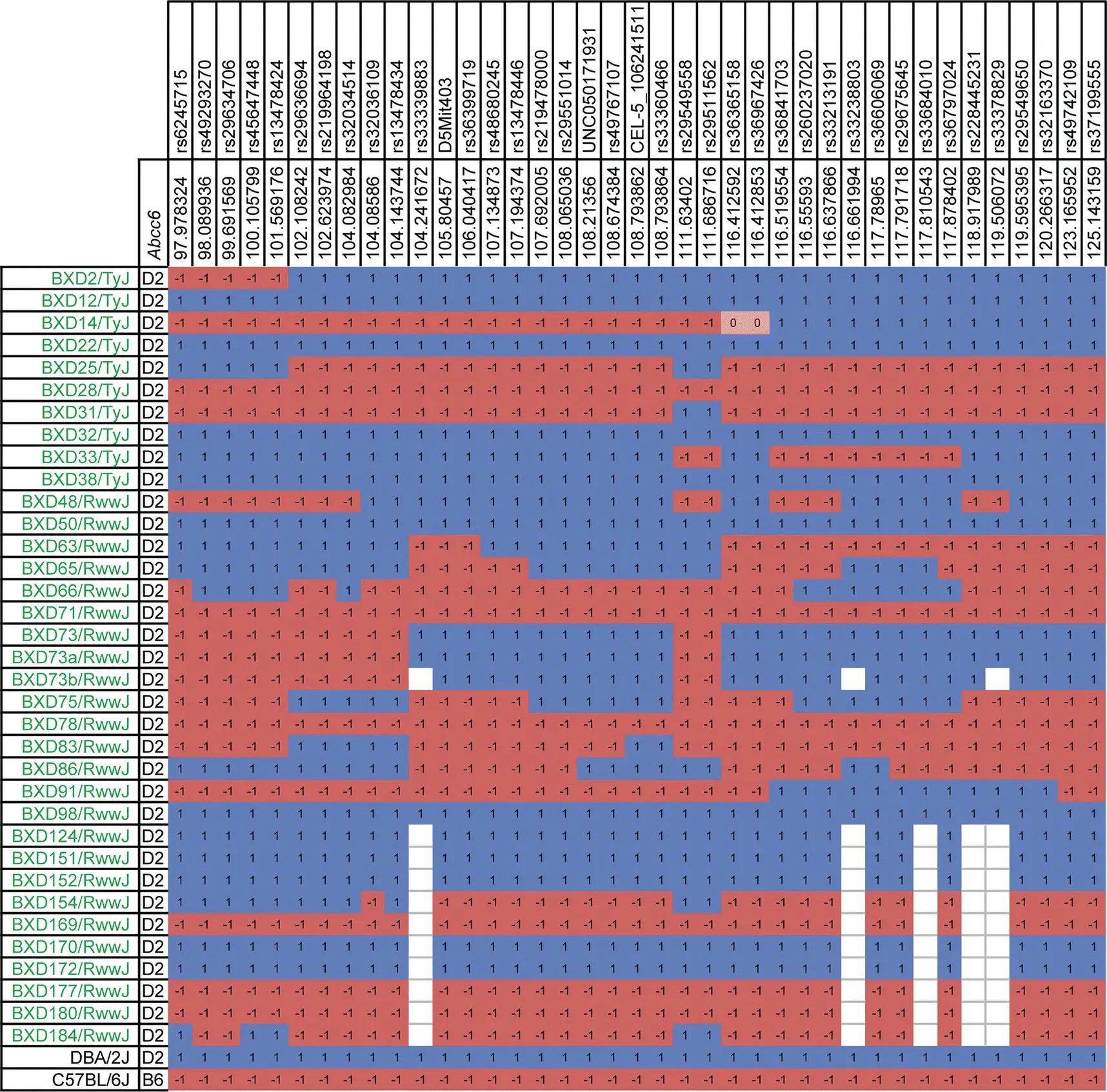
Figure 3.7 Using recombinant inbred lines to narrow candidate gene intervals using WWW.GeneNetwork.org The BXD lines on the left are all homozygous for the hypomorphic Abcc6 allele from DBA/2J. One quantitative trait locus interval has a mixture of DBA/2J (blue) and C57BL/6J (red) DNA. By phenotyping the mice, it is possible to narrow the interval to identify the modifier gene.
Source: Generated by Dr. Jason Bubier 4 May 2020; WWW.GeneNetwork.org.
Nomenclature for congenic strains begins with a standard abbreviation for the host strain ( Table 3.4), followed by a period (.) to indicate congenicity, then the standard abbreviation for the donor strain, then a hyphen, then the allele(s) or congenic interval transferred, then a forward slash (/), and then laboratory code of the investigator in whose laboratory the congenic was completed. For example, a mutation in the sodium channel, voltage‐gated, type VIII, alpha gene ( Scn8a 8J) was induced using N ‐ethyl‐ N ‐nitrosourea (ENU) in the C57BL/6J background and was backcrossed onto C3HeB/FeJ in the laboratory of Dr. Wayne Frankel so the strain name is C3Fe.B6‐ Scn8a 8J/Frk [24]. In the strain B6.C3‐ Pde6b rd1 Hps4 le/J both the phosphodiesterase 6B, cGMP, rod receptor, β‐polypeptide retinal degeneration one allele ( Pde6b rd1) and HPS4, biogenesis of lysosomal organelles complex 3 subunit 2 light‐ear allele ( Hps4 le) were transferred in a single congenic interval from Chromosome 5 of a mutant subline of C3H/HeJ onto C57BL/6J. The congenic interval can also be represented not by the allele(s) of interest but by the most proximal and distal markers identified in the donor interval, such as in the strain B6.129S1‐(rs13480546‐rs13480629)/Kjn [25]. If two alleles of interest from separate genetic origins are combined into one congenic strain or if the precise origins of the donor sequence are not known, then there is no single donor strain that can be put after the period so that complex genomic donor source is represented as “Cg.” The Tg(Ins2‐rtTA)2Doi transgenic mice were generated by microinjection into (C57BL/6J x SJL/J)F1 hybrid mice then backcrossed onto a NOD host to generate the congenic named NOD.Cg‐Tg(Ins2‐rtTA)2Doi/Doi [26], in which the Cg is used because it is unknown whether the congenic interval derives from C57BL/6J or SJL/J. If the source of the desired donor sequence does not come from a strain that is a pure inbred, but rather contains contributions from a source different from the host background or the donor congenic interval, then that strain information can be represented in parentheses after the donor strain abbreviation. Thus, if it were known that Tg(Ins2‐rtTA)2Doi inserted into C57BL/6J‐derived sequence in the original F1 hybrid, and not into SJL/J‐derived sequence, then the strain name of the congenic would be NOD.B6(SJL)‐Tg(Ins2‐rtTA)2Doi/Doi. This allows the reader to see at a glance that there is a possibility that some small amounts of SJL/J‐derived sequence exist in the strain. In the example of NOD.129S4(B6)‐ Art2a tm1Fkn Art2b tm1Fkn/Lt it is immediately clear that both targeted alleles were generated in 129S4, at least one, if not both, were bred at least once to C57BL/6J, and then both were backcrossed onto NOD, either together or separately then bred together [27]. If there are more than one additional sources of trace contribution to the genetic background, aside from the donor, then Cg is entered into the parentheses to represent this. For example, NOD.129P2(Cg)‐ Il10 tm1Cgn Il4 tm1Cgn/Dvs had C57BL/6J and C57BL/10J in part of the genetic lineage so the potential contributions from those backgrounds are captured in the (Cg) [28]. Another example of a congenic strain in which the donor origin is represented by “Cg” is B6.Cg‐ Kitl Sl Krt71 Ca/J, a C57BL/6J host congenic for both Kitl Sl, which arose in C3H/He, and Krt71 Ca, which arose in a Swiss stock [29].
Table 3.9 Congenic strains.
| Advantages Genetic and phenotypic uniformityReduce experimental variabilityCan transfer most mutations onto a different genetic backgroundAllows examination of modifier genesCan maintain mutation or transgene homozygously and use inbred as control |
| Disadvantages Generation time (two to three years to N10 or longer if ovarian graft to immunodeficient mouse used; can reduce time with speed congenics)Phenotype may change on different genetic backgroundLinked genes may confuse experimental findings |
As illustrated in the above examples, more than one gene can be selected for in the congenic breeding process, especially if molecular markers are available so that each generation can be screened. The entire process is speeded up if markers for the host strain are identified on all chromosomes and selected for during the inbreeding process, to create what are termed speed congenics.
Читать дальшеИнтервал:
Закладка:
Похожие книги на «Pathology of Genetically Engineered and Other Mutant Mice»
Представляем Вашему вниманию похожие книги на «Pathology of Genetically Engineered and Other Mutant Mice» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Pathology of Genetically Engineered and Other Mutant Mice» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.












