 0
0
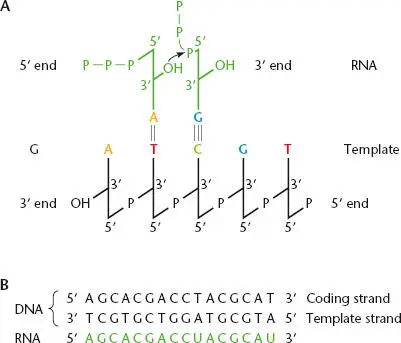
Figure 2.5 RNA transcription. (A)The polymerization reaction, in which incoming nucleoside triphosphates (NTPs) pair with the template strand of DNA during transcription and are joined to generate the RNA chain. The β and γ phosphates of each incoming NTP (other than the initiator NTP) are removed as pyrophosphate (PP i). (B)The coding strand (or nontemplate strand) of the DNA has the same sequence as the mRNA (with T residues in the DNA replaced with U residues in the RNA). The template strand is the DNA strand to which the mRNA is complementary.
RNA transcripts are copied only from selected regions of the DNA, rather than from the whole molecule; therefore, the RNA polymerase holoenzyme can start making an RNA chain from a double-stranded DNA only at certain sites. These DNA regions are called promoters, and the RNA polymerase recognizes a particular nucleotide (usually a T or C) in the promoter region of the template strand as a transcription start site, shown as +1 in Figure 2.6. Thus, the first base in the chain is usually an A or a G laid down opposite to a T or C, respectively.
The RNA polymerase holoenzyme recognizes different types of promoters on the basis of which type of σ factor it contains. The most common promoters are those recognized by the RNA polymerase with the σ called σ 70in E. coli . The σ factors are often named for their size, and this one has a molecular mass of 70,000 Da (70 kDa). Replacement of σ 70with a different σ factor results in an RNA polymerase holoenzyme that recognizes a different set of promoters; this will be discussed in later chapters on gene regulation.
Promoters recognized by holoenzymes containing the same σ are not identical to each other, but they do share certain sequences, known as consensus sequences, by which they can be distinguished. Figure 2.6shows the consensus sequence of promoters recognized by holoenzymes containing σ 70in E. coli , which illustrates a common pattern for promoter structure . The promoter sequence has two important regions: a short AT-rich region centered about 10 bp upstream of the transcription start site, known as the – 10 sequence, and a second region centered about 35 bp upstream of the start site, called the –35 sequence. The σ 70factor usually must bind to both sequences to start transcription (see below) but does not require that the DNA have a perfect match to these consensus sequences; binding to the promoter occurs only when σ 70is in the holoenzyme complex. Sequence-specific binding to the promoter determines not only the site at which transcription will initiate, but also the direction the RNA poiymerase will move along the DNA (in other words, which strand of the DNA will be transcribed from a given region).
THE STEPS OF TRANSCRIPTION
Figure 2.7shows an overview of the steps of transcription. The RNA polymerase holoenzyme recognizes a promoter and begins transcription with a ribonucleoside triphosphate. As the RNA chain begins to grow, the RNA polymerase holoenzyme releases its σ factor, and the five-subunit core enzyme continues to move along the template DNA strand in the 3′-to-5′ direction, synthesizing RNA in the 5′-to-3′ direction. Inside an opening in the DNA helix approximately 17 bases long, called the transcription bubble, the elongating RNA and the template DNA strand pair with each other to form a DNA-RNA hybrid of approximately 8 or 9 bp with a double-helix structure similar to that of a double-stranded DNA molecule. As RNA polymerase moves along the DNA, the upstream portion of the DNA-RNA helix separates as new ribonucleotides are incorporated into the 3′ end of the growing RNA chain, and the 8-to 9-bp hybrid is maintained. The resulting RNA product emerges from RNA polymerase through a channel, and the DNA strands behind the RNA polymerase rebind to each other. The RNA polymerase continues to move along the DNA template until it reaches a terminator, which signals the RNA polymerase to release both the DNA template and the RNA transcript.
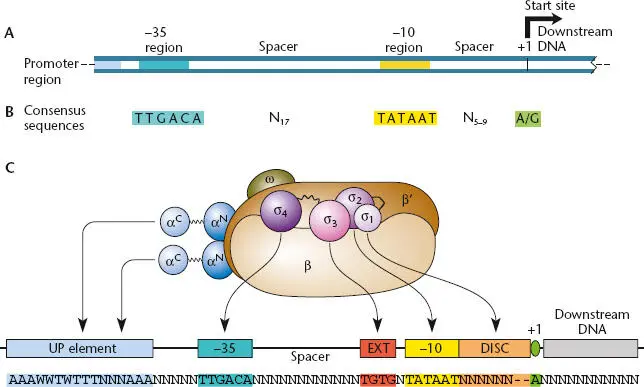
Figure 2.6 (A)Typical structure of a σ 70bacterial promoter. (B)The consensus sequences of a σ 70bacterial promoter. RNA synthesis typically starts with an A or a G, and no primer is required. N, any nucleotide. (C)Positions of interaction between RNA polymerase and promoter DNA.
It was once assumed that after initiation occurs, the RNA polymerase moves along the DNA at a uniform rate, polymerizing nucleotides into RNA. However, it is now known that the RNA polymerase often starts making RNA and then repeatedly aborts, synthesizing a number of short RNAs in a process called abortive initiationbefore finally leaving the promoter. Even after transcription is under way, RNA polymerase often pauses and sometimes even backs up before continuing. In this section, we discuss in more detail each of the steps in transcription ( Figure 2.8), which have been established over many years by a large number of researchers. We discuss these steps one at a time because each of them is the basis for regulatory mechanisms that are discussed in later chapters.
In the first step ( Figure 2.9), the RNA polymerase core enzyme binds to a σ factor to form the holoenzyme. The bound σ factor then directs the complex to the correct promoter in a process called promoter recognitionor binding. The σ factor must be able to recognize the promoter even though the DNA in the promoter is still in a double-stranded state. Sigma factors consist of a number of domains held together by flexible linkers. Most σ factors are related to σ 70, and their domains play similar roles in recognizing their specific promoters. Figure 2.6Cshows the conserved regions of the σ 70family of sigma factors and the roles played by some of the conserved domains in promoter recognition and initiation of transcription.
One domain of the bound σ, σ 4, recognizes the –35 sequence when it is still in the double-stranded state. Another σ domain, σ 2, binds to the AT-rich –10 sequence. Some σ 70promoters have additional sequence elements that interact with other domains of the RNA polymerase ( Figures 2.6and 2.10). The efficiency of binding of RNA polymerase to a promoter can be enhanced by sequences upstream of the promoter, called UP (for upstream) elements, to which the carboxy terminus of the α subunits, called αCTD (for α subunit carboxyl-terminal domain), can bind and help stabilize the binding of RNA polymerase to the DNA. A flexible domain that links αCTD and the amino-terminal domain of α (αNTD) allows αCTD to reach the UP element on the DNA. Also, some promoters have what is called an extended –10 sequence (TGN, located immediately upstream of the –10 sequence to give the sequence TGNTATAAT). This sequence is recognized by the σ 3domain and is often found in promoters that lack a –35 sequence that is efficiently recognized by σ 4. The similarity of a promoter sequence to the consensus sequences for a particular σ, in combination with other elements that interact with other domains of RNA polymerase, dictates the efficiency with which a promoter is recognized by holoenzyme containing that σ.
Читать дальше

 0
0










