Figure 1.16shows how the one-way nature of ter sequences can cause replication to terminate in a specific region of the chromosome. In the illustration, two ter sites called terA and terB bracket the termination region. Replication forks are unaffected by the terA site in the clockwise direction but are terminated in the counterclockwise direction. The opposite is true for terB . Thus, the clockwise-moving replication fork progresses through terA , but if it gets to terB before it meets the counterclockwise-moving fork, it stalls, because it cannot move clockwise through terB . Similarly, the replication fork moving in the counterclockwise direction stalls at the terA site and waits for the clockwise-moving replication fork. When the counterclockwise and clockwise replication forks meet, at terA , terB , or somewhere between them, the two forks terminate replication, releasing the two daughter DNAs. In E. coli , it is known that most DNA replication termination occurs at one ter site, possibly because it is oriented to terminate replication forks traveling in the clockwise direction, which is shorter in most laboratory E. coli strains.
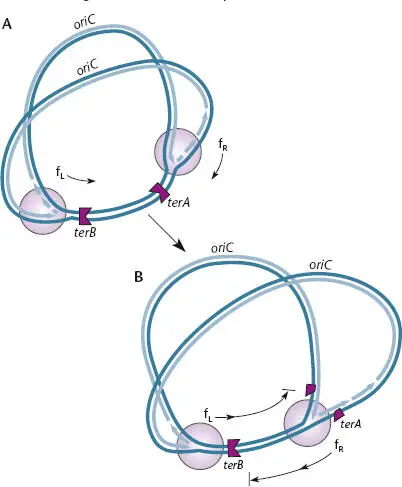
Figure 1.16 Termination of chromosome replication in Escherichia coli . (A)The replication forks that start at the origin of chromosomal replication ( oriC ) can traverse terA and terB in only one direction, opposite that indicated by the black arrows. (B)When they meet, between or at one of the two clusters, chromosome replication terminates. f Lis the fork that initiated to the left and moved in a counterclockwise direction. f Ris the fork that initiated to the right and moved in a clockwise direction. Adapted from Camara JE, Crooke E, in Higgins MP, ed, The Bacterial Chromosome (ASM Press, Washington, DC, 2005), with permission.
Encountering a ter DNA sequence, by itself, is not sufficient to stop the replication fork. A protein is also required to terminate replication at ter sites. The protein that works with ter sites, called the terminus utilization substance (Tus) in E. coli and the replication terminator protein (RTP) in B. subtilis , binds to the ter sites and stops the replicating helicase (DnaB in E. coli ) that is separating the strands of DNA ahead of the replication fork. In both E. coli and B. subtilis , multiple ter sites bracket the terminus region, helping to ensure that replication proceeds from oriC to the terminus region (see Box 1.1). While the ter systems are not absolutely essential for E. coli and B. subtilis growing in the laboratory setting, they are important for other aspects of genome stability that are important for maintaining genome integrity over time in the natural environment (see Rudolph et al., Suggested Reading). It has been argued that the active termination systems involving ter sites and a trans acting protein originated in plasmids ( chapter 5) and were domesticated in some branches of bacteria for use in the bacterial chromosome (see Galli et al., Suggested Reading).
BOX 1.1
Structural Features of Bacterial Genomes
It is widely appreciated that the chromosomes are the information storehouse for an organism. What is less appreciated is that the chromosome as a structure has evolved sequence motifs that allow it to be efficiently replicated, repaired, and segregated into daughter cells. The distribution and orientation of these motifs are discussed here; the molecular biology of the systems that recognize these sequences is explained in greater detail in the text. The placement of these sequence motifs in the context of the chromosome is important for their function, as is the orientation of many of these sequences. Many of the motifs are oriented in one direction, which follows the direction of the DNA replication fork. DNA replication in E. coli and B. subtilis (and all bacteria studied to date) is initiated within a single oriC region and continues bidirectionally to a position on the chromosome equidistant from the origin (indicated by the long arrow-headed line in the figure). The dif site where the resolution of dimer chromosomes occurs is found near to where DNA replication normally terminates.
Certain DNA sequence motifs that guide DNA replication and DNA repair are polar in that they are not symmetrical and need to be in a specific 5′-to-3′ direction to carry out their functions. In other words, these sequences must be found in a certain orientation in the chromosome and will not work if they are flipped around. In E. coli and B. subtilis , DNA replication forks are actively terminated at specific sites called ter sites. The ter sites act as a trap for DNA replication forks, and these sites encompass a large portion of the chromosome, allowing DNA replication forks to pass when approaching from one direction but not the other. Multiple ter sites (10 in E. coli and 9 in B. subtilis ) are found in the genome, and the redundancy of these sites may be important for catching replication events that get through the initial ter sites (indicated with a dashed line) or to stop replication forks that are initiated for DNA repair by recombination (see chapter 9). For unknown reasons, the central ter sites in B. subtilis are very close together, while the central sites in E. coli are separated by hundreds of thousands of base pairs. Along the path of DNA replication are sequence motifs involved in guiding the DNA translocase proteins involved in chromosome segregation: FtsK, found in E. coli (which recognizes motifs called KOPS), and SpoIIIE, found in B. subtilis (called SRS motifs). The StpA DNA translocase from B. subtilis may also recognize the SRS sites. Chromosomes also have polar DNA sequences that guide the recombination machinery (see chapter 9). DNA recombination is extremely important in bacteria as a way to repair DNA double-strand breaks that occur during DNA replication. Repair of these breaks occurs when recombination reestablishes a DNA replication fork using one broken end and the sister chromosome. Reestablishment of DNA replication involves an efficient processing event that utilizes polar sites called chi sites. The RecBCD complex in E. coli or the AddAB complex in B. subtilis carries out this processing activity using information found in the chi sites . chi sites are species specific and are common in genomes in one orientation from the origin to the terminus region on the leading strand (found about 1 every 5 kb in the E. coli chromosome).
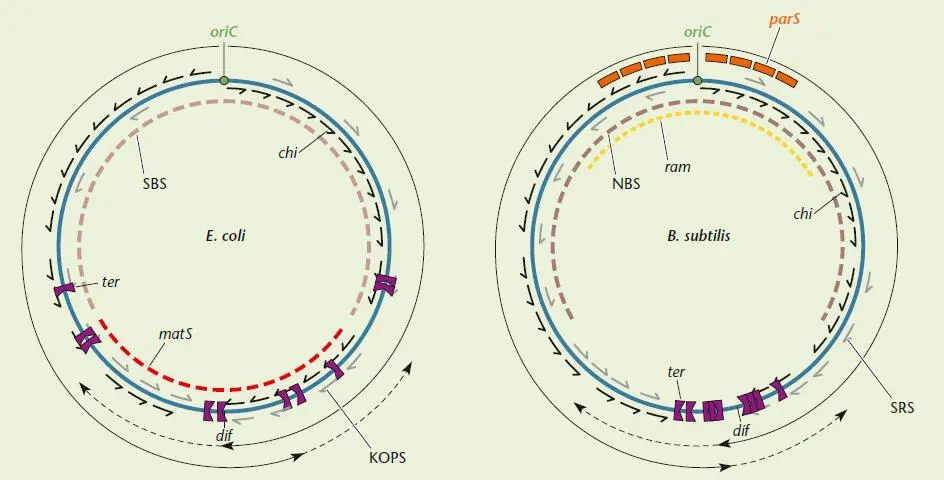
Other polar sequence biases in the chromosome include an overrepresentation of the 5′-CTG-3′ sequence that primes lagging-strand DNA synthesis (not shown). Interestingly, the most common triplet codon is the CUG (5′-CTG-3′ in DNA) that codes for leucine, comprising almost 5% of all codons in E. coli . The 5′-CTG-3′ sequence is found in the chi sequence and all of the most frequent 8-bp sequences in the chromosome. It would be difficult to argue which came first, the use of this sequence by the primase or its frequency of use as a codon. Another type of sequence bias, but one that is not polar in nature, is a general sequence bias called the G/C skew, where G and C are overrepresented in the leading strand. The trend toward A and T in the lagging strand is believed not to have an adaptive value but to be a result of the way in which repair differs on the two strands.
Читать дальше













