Genetic Analysis of Complex Disease
Здесь есть возможность читать онлайн «Genetic Analysis of Complex Disease» — ознакомительный отрывок электронной книги совершенно бесплатно, а после прочтения отрывка купить полную версию. В некоторых случаях можно слушать аудио, скачать через торрент в формате fb2 и присутствует краткое содержание. Жанр: unrecognised, на английском языке. Описание произведения, (предисловие) а так же отзывы посетителей доступны на портале библиотеки ЛибКат.
- Название:Genetic Analysis of Complex Disease
- Автор:
- Жанр:
- Год:неизвестен
- ISBN:нет данных
- Рейтинг книги:4 / 5. Голосов: 1
-
Избранное:Добавить в избранное
- Отзывы:
-
Ваша оценка:
- 80
- 1
- 2
- 3
- 4
- 5
Genetic Analysis of Complex Disease: краткое содержание, описание и аннотация
Предлагаем к чтению аннотацию, описание, краткое содержание или предисловие (зависит от того, что написал сам автор книги «Genetic Analysis of Complex Disease»). Если вы не нашли необходимую информацию о книге — напишите в комментариях, мы постараемся отыскать её.
An up-to-date and complete treatment of the strategies, designs and analysis methods for studying complex genetic disease in human beings Genetic Analysis of Complex Diseases
Genetic Analysis of Complex Diseases
Genetic Analysis of Complex Diseases
Genetic Analysis of Complex Disease — читать онлайн ознакомительный отрывок
Ниже представлен текст книги, разбитый по страницам. Система сохранения места последней прочитанной страницы, позволяет с удобством читать онлайн бесплатно книгу «Genetic Analysis of Complex Disease», без необходимости каждый раз заново искать на чём Вы остановились. Поставьте закладку, и сможете в любой момент перейти на страницу, на которой закончили чтение.
Интервал:
Закладка:
Ascertainment Bias
In genetic studies, research subjects are selected for participation based on the presence or absence of the trait of interest. The family member who comes to the investigator’s attention (through admission to a hospital or solicitation of support groups, for example) is called the proband. Most often, the proband is an individual who exhibits the trait of interest. Ascertainment through an affected individual can lead to a bias in the distribution of the numbers of affected and unaffected family members present in the analysis. Because the ascertainment scheme necessitated that the family have at least one affected individual (proband), families that may be carrying the genetic liability of interest but, by chance, do not contain an affected family member will not be ascertained. This phenomenon is referred to as ascertainment bias and is demonstrated in Figure 3.2. Depending on the analysis, ascertainment bias may greatly influence the outcome of the analyses.
In general, ascertainment bias should not affect the ability to accept or reject linkage in linkage analysis ( Chapter 6). However, it can affect the estimate of the recombination fraction between the genetic marker and the disease locus (Vieland and Hodge 1996). Ascertainment bias can also influence familial recurrence risk ratios ( λ R) (Guo 1998; Cordell and Olson 2000) and the estimate of the segregation probability of the disease locus in segregation analysis (Ewens and Shute 1986; Greenberg 1986; Stene 1989). Furthermore, it has been argued that in some cases, ascertainment bias may be a reasonable explanation for what appears to be genetic anticipation in some pedigrees (Penrose 1948).
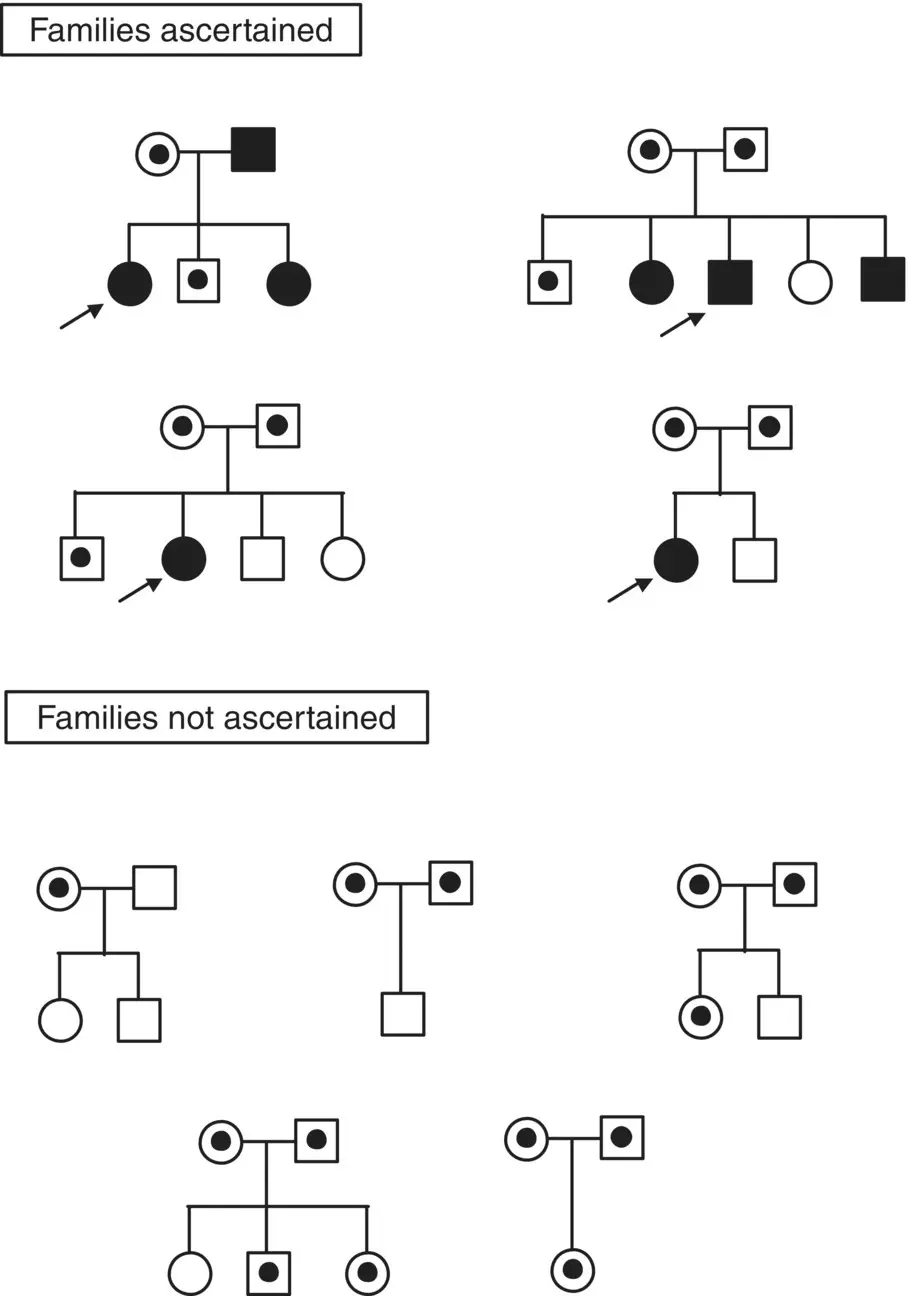
Figure 3.2 Example of ascertainment bias in genetic analysis when ascertaining through an affected individual.
Clearly, ignoring the ascertainment scheme used in an analysis can lead to false conclusions. For example, in the fragile X literature, it had been reported that male offspring of premutation and full‐mutation mothers received larger CGG repeat expansions than did female offspring (Rousseau et al. 1994; Loesch et al. 1995). However, once potential sources of ascertainment bias were removed, it was determined that there was no association between sex of the offspring and the size of the mutation inherited from the mother (Ashley‐Koch et al. 1998). In this example, there were three types of ascertainment bias that were sequentially removed from the analysis. First, in clinically ascertained families, the transmission to the proband was removed. This is a standard ascertainment correction known as the Weinberg correction scheme (Weinberg 1912). Second, transmissions were excluded where not all individuals within a sibship were tested. That is, in fragile X families, nonsymptomatic females are tested more often than nonsymptomatic males because only the females are at risk to have affected offspring. This ascertainment bias increases the number of transmissions to premutation females compared with premutation males. Finally, the transmissions of the CGG repeat to the mothers of the proband were omitted. Because full‐mutation females have reduced fitness (Sherman et al. 1984), mothers of probands are more likely to be premutation carriers merely because they have reproduced. Consequently, including transmissions of the CGG repeat from carrier grandmothers to mothers of probands will also increase the number of transmissions to premutation females. Table 3.1shows how sequentially correcting for the ascertainment bias in fragile X families changes the conclusion regarding the association between offspring sex and the size of the CGG expansion. Notice that the statistical tests (t‐test and logistic regression) become insignificant as the correction schemes are applied. Additionally, it illustrates that in some cases, standard correction schemes, such as the Weinberg method, are not sufficient to abolish the bias in the ascertainment scheme. Thus, all potential sources of bias in the data set must be carefully considered.
Table 3.1 Association between sex of offspring and risk of expansion in fragile X syndrome.
Source: From Ashley‐Koch et al. (1998); reprinted with permission.
| Proportion of offspring with full mutation (%) | P ‐value | |||
|---|---|---|---|---|
| Ascertainment scheme (no. of cases) | Males | Females | t‐test | Logistic regression |
| Removal of cases associated with ascertainment (434) | 0.46 | 0.38 | 0.06 | 0.07 |
| Removal of incompletely ascertained sibships (338) | 0.48 | 0.43 | 0.38 | 0.34 |
| Removal of transmissions to proband’s mother (298) | 0.48 | 0.50 | 0.63 | 0.71 |
Approaches to Determining the Genetic Component of a Disease
King et al (1984) described the steps necessary to define the genetic mechanisms involved in a disease or trait many years ago, but those steps are still valid today. First, the evidence for a familial component to the condition must be established. Next, the cause of familial aggregation must be determined. That is, clustering in a disease may result from common environmental factors, rather than genetic factors, and these two hypotheses must be evaluated. Finally, the specific genetic factors must be identified and the manner in which they interact with each other and with environmental factors to contribute to the disease etiology must be defined.
Below, several approaches are presented to evaluate whether or not genes contribute to the etiology of a disease, and to quantitate the contribution of those genes to the disease etiology.
Co‐segregation with Chromosomal Abnormalities and Other Genetic Disorders
While complex disorders generally do not exhibit a recognizable inheritance pattern, occasionally in a subset of patients, a complex disorder will segregate with a cytogenetic abnormality or another known genetic disorder. These associations provide valuable information regarding the location of at least one locus involved in the disease etiology. For example, individuals with trisomy 21, or Down syndrome, have an increased risk for developing Alzheimer disease. This increased risk is due to amyloid plaques resulting from an increased dosage of the amyloid precursor protein (APP) (Rumble et al. 1989). Thus, it was not surprising to find that a subset of families with early‐onset Alzheimer disease are linked to chromosome 21 and segregate mutations in the APP gene (St George‐Hyslop et al. 1987; Goate et al. 1991). Another example of co‐segregation of a complex disease and cytogenetic abnormality is the association between autistic disorder and 15q11‐q13 abnormalities. There are numerous examples of isolated patients with autistic disorder and duplications or inversions involving 15q11‐q13 (Wolpert et al. 2000). In many cases, the de novo rearrangements are thought to be maternal in origin (Lindgren et al. 1996). In addition, in the absence of these cytogenetic abnormalities, families with two or more individuals exhibiting autistic disorder display evidence for linkage (Philippe et al. 1999; Bass et al. 2000) and linkage disequilibrium (Cook, Jr. et al. 1998; Martin et al. 2000a; Menold et al. 2000) in this region. Although as yet no gene has been identified, the convergence of cytogenetic, linkage, and association data suggest that a locus involved in susceptibility to autistic disorder is located at 15q11‐q13.
Familial Aggregation
Интервал:
Закладка:
Похожие книги на «Genetic Analysis of Complex Disease»
Представляем Вашему вниманию похожие книги на «Genetic Analysis of Complex Disease» списком для выбора. Мы отобрали схожую по названию и смыслу литературу в надежде предоставить читателям больше вариантов отыскать новые, интересные, ещё непрочитанные произведения.
Обсуждение, отзывы о книге «Genetic Analysis of Complex Disease» и просто собственные мнения читателей. Оставьте ваши комментарии, напишите, что Вы думаете о произведении, его смысле или главных героях. Укажите что конкретно понравилось, а что нет, и почему Вы так считаете.











