The FIS protein plays a key role in controlling the expression of the dps gene during the growth cycle (Grainger et al. 2008). This gene is transcribed from a single promoter that is recognised by both the RpoD‐ and the RpoS‐containing forms of RNA polymerase (Altuvia et al. 1994). Three NAPs are involved in the regulation of dps transcription: FIS, H‐NS, and IHF. IHF and the OxyR transcription factor activate the dps promoter in association with RpoD in bacteria experiencing oxidative stress; in stationary phase the same promoter is utilised by RNA polymerase containing RpoS (Altuvia et al. 1994). During exponential growth, FIS holds RNA polymerase containing RpoD at the promoter while H‐NS excludes the RpoD form of the RNA polymerase holoenzyme from the same promoter. The transcriptionally inert FIS‐RpoD‐RNA‐polymerase complex prevents entry of the RpoS‐containing RNA polymerase holoenzyme. Since FIS levels decline to negligible values as the bacterium enters stationary phase, this negative control is no longer exerted, and as H‐NS is unable to impede the activity of the RpoS‐containing form of RNA polymerase, dps transcription can begin (Grainger et al. 2008).
1.42 The H‐NS Protein: A Silencer of Transcription
Throughout the 1980s, the gene that encodes the H‐NS NAP was discovered and rediscovered by investigators working independently of each other because this protein is involved in controlling the expression of so many different components of the bacterium. One of the consequences of the broad influence of H‐NS is that the gene that encodes it has been given many names, such as bglY , osmZ , pilG , virR , and dxdR , among others. In each case, the name linked a mutation in the gene, now referred to universally as hns , to a specific H‐NS‐dependent system in the cell such as beta‐glucoside uptake and utilisation ( bglY ), pilus expression ( pilG ), the osmotic stress response ( osmZ ), expression of a virulent phenotype in the pathogen Shigella ( virR ), or thermo‐regulated adhesin expression in E. coli ( drdX ) (Defez and de Felice 1981; Higgins et al. 1988; Göransson et al. 1990; Maurelli and Sansonetti 1988; Spears et al. 1986). The hns gene is located in the Ter macrodomain of the chromosome, close to the topA gene, and early experiments revealed a connection between some hns alleles and alterations in the DNA topology of reporter plasmids in those strains (Dorman et al. 1990; Higgins et al. 1988). The finding that H‐NS binding to DNA impedes access to that DNA by DNA gyrase may explain these observations (Sutormin et al. 2019). H‐NS can influence transposition and site‐specific recombination as well as transcription (Corcoran and Dorman 2009; Liu et al. 2011; Whitfield et al. 2009) and it has effects at the level of mRNA translation too (Park et al. 2010).
The H‐NS protein is a small, abundant NAP that is produced at all stages of the growth cycle (Ali Azam and Ishihama 1999; Dorman 2013; Free and Dorman 1995). It binds to A+T‐rich DNA and has been described as having a preference for DNA with intrinsic curvature (Yamada et al. 1991). These features are commonly associated with transcriptional promoters, allowing H‐NS to target large numbers of genes for repression, even if the functions of the gene products are not related in any obvious way. The protein is almost always a repressor, allowing it to silence transcription of a large subset of the genes in the genome ( Figure 1.21).
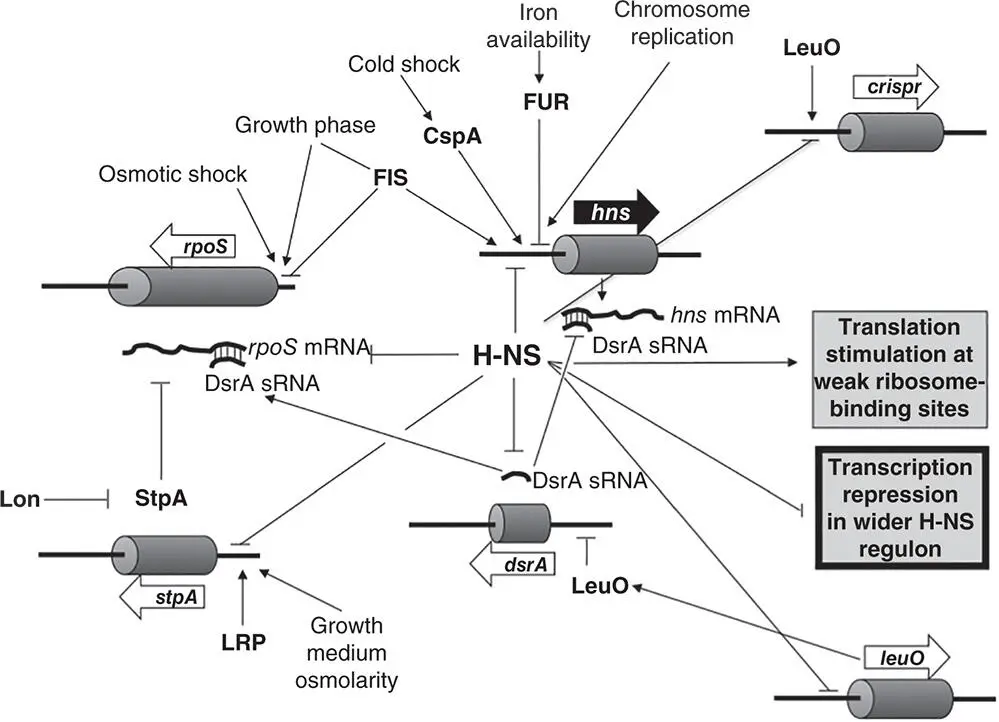
Figure 1.21The vast H‐NS regulon. The H‐NS protein controls the expression of hundreds of genes in the pan genome, the accessory genome, and the core genome. Its own gene is also subject to complex control at the transcriptional and posttranscriptional levels. Among the factors influencing hns transcription positively are chromosome replication, the growth‐phase‐dependent FIS protein, and the cold‐shock regulatory protein CspA; it is auto‐repressed by its own gene product and repressed by the iron‐binding Fur protein, and translation of H‐NS's own mRNA is inhibited by the DsrA sRNA in an Hfq‐dependent manner. H‐NS, in turn, inhibits DsrA expression, resulting in a positive effect throughout the DsrA regulon, including production of the RpoS sigma factor. H‐NS silences crispr transcription, an effect that is antagonised by the LeuO transcription factor, with the leuO gene in turn being H‐NS‐repressed. Like StpA (whose gene H‐NS represses) the H‐NS protein can act at the level of RNA, for example improving the translation of the maltose regulatory gene by repositioning ribosomes so that translation can proceed. The H‐NS paralogue, StpA, is a substrate for Lon‐mediated proteolysis but dimerisation with H‐NS protects StpA from this fate (H‐NS is not degraded by Lon). StpA can act as a backup for H‐NS and its expression is governed by an independent set of cues, such as the LRP protein and bacterial growth phase (H‐NS is present at all stages of growth). The H‐NS regulon includes genes involved in bacterial virulence and whose expression is controlled by pH, osmotic stress, temperature, and other environmental influences. Its (usually negative) influence is overcome by an impressive array of mechanisms that link the expression of the H‐NS target genes to information relevant to the infection process (see Stoebel et al. 2008b). Arrows represent positive regulatory inputs and negative ones are indicated by ‘T’ symbols.
The very low base sequence requirement, as opposed to base content requirement, of H‐NS for its binding sites allows this protein to interact with DNA on the basis of DNA shape rather than nucleotide sequence. Its reliance on indirect readout for DNA‐binding‐site recognition underlies the contribution made by H‐NS to silencing the transcription of genes that have been acquired by HGT in Gram‐negative organisms (Dorman 2007, 2014a). This xenogeneic silencing is hypothesised to allow potentially harmful genes to be imported into the genome without compromising the competitive fitness of the bacterium (Dorman 2004, 2007; Lucchini et al. 2006; Navarre et al. 2006; Oshima et al. 2006). It should be pointed out that although H‐NS displays low nucleotide sequence specificity at its binding sites, a preferred sequence has been identified at the proU operon (Bouffartigues et al. 2007; Lang et al. 2007). This operon encodes a transport system for glycine‐betaine, a modified amino acid that protects macromolecules in bacteria from the deleterious effects of water loss following osmotic shock (Cairney et al. 1985). The proU promoter is induced by osmotic up‐shock in the presence of potassium ions and is subject to repression by the H‐NS protein (Gowrishankar and Manna 1996; Higgins et al. 1988; Sutherland et al. 1986). It contains two matches to an A+T‐rich consensus sequence that contains the very flexible TpA di‐nucleotide step, possibly important in specifying local DNA curvature (Bouffartigues et al. 2007; Lang et al. 2007). This motif may represent an evolutionary step towards a more specific binding site for the H‐NS protein.
H‐NS forms dimers and these can link together to form higher‐order oligomers (Arold et al. 2010; Shahul Hameed et al. 2018) ( Figure 1.22). The monomer consists of an amino‐terminal dimerisation domain that is connected by a linker region to a carboxyl‐terminal nucleic-acid‐binding domain (Dorman et al. 1999). The dimer can bridge different parts of the same DNA molecule or link separate DNA molecules together, activities that have been confirmed in vitro using single molecule methods and atomic force microscopy (Dame et al. 2000, 2005, 2006). The protein displays two DNA‐binding modes, one involving bridging of two DNA duplexes (or two sections of the same DNA duplex) and the other involving polymerisation of H‐NS along a single DNA duplex ( Figure 1.23) (Corcoran and Dorman 2009; O'Gara and Dorman 2000; Lim et al. 2012). Polymerisation, with or without bridging, occurs following initial binding of H‐NS to a preferred nucleation site (Badaut et al. 2002). In vitro data indicate that changes in the concentration of magnesium ions can toggle H‐NS between these two binding modes (Liu et al. 2010), though the significance of this effect in the bacterial cell is unknown. In addition to the influence of divalent cations, the toggling of H‐NS between these two binding modes is modulated by co‐regulator proteins such as Hha, YdgT, and YmoA, which resemble the dimerisation domain of H‐NS (van der Valk et al. 2017). By coating the DNA surface, H‐NS can exclude other proteins or protein complexes, including RNA polymerase ( Figure 1.23). However, the negative influence of H‐NS on transcription can be overcome in a variety of ways (Dorman and Kane 2009; Stoebel et al. 2008b).
Читать дальше